Small Gtpases* 1,2 3§ David J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

G Protein Alpha 13 (GNA13) (NM 006572) Human Tagged ORF Clone Lentiviral Particle Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC207762L3V G protein alpha 13 (GNA13) (NM_006572) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: G protein alpha 13 (GNA13) (NM_006572) Human Tagged ORF Clone Lentiviral Particle Symbol: GNA13 Synonyms: G13 Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) ACCN: NM_006572 ORF Size: 1131 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC207762). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_006572.3 RefSeq Size: 4744 bp RefSeq ORF: 1134 bp Locus ID: 10672 UniProt ID: Q14344, A0A024R8M0 Domains: G-alpha Protein Families: Druggable Genome Protein Pathways: Long-term depression, Regulation of actin cytoskeleton, Vascular smooth muscle contraction MW: 44 kDa This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 G protein alpha 13 (GNA13) (NM_006572) Human Tagged ORF Clone Lentiviral Particle – RC207762L3V Gene Summary: Guanine nucleotide-binding proteins (G proteins) are involved as modulators or transducers in various transmembrane signaling systems (PubMed:15240885, PubMed:16787920, PubMed:16705036, PubMed:27084452). -

Ciliopathiesneuromuscularciliopathies Disorders Disorders Ciliopathiesciliopathies
NeuromuscularCiliopathiesNeuromuscularCiliopathies Disorders Disorders CiliopathiesCiliopathies AboutAbout EGL EGL Genet Geneticsics EGLEGL Genetics Genetics specializes specializes in ingenetic genetic diagnostic diagnostic testing, testing, with with ne nearlyarly 50 50 years years of of clinical clinical experience experience and and board-certified board-certified labor laboratoryatory directorsdirectors and and genetic genetic counselors counselors reporting reporting out out cases. cases. EGL EGL Genet Geneticsics offers offers a combineda combined 1000 1000 molecular molecular genetics, genetics, biochemical biochemical genetics,genetics, and and cytogenetics cytogenetics tests tests under under one one roof roof and and custom custom test testinging for for all all medically medically relevant relevant genes, genes, for for domestic domestic andand international international clients. clients. EquallyEqually important important to to improving improving patient patient care care through through quality quality genetic genetic testing testing is is the the contribution contribution EGL EGL Genetics Genetics makes makes back back to to thethe scientific scientific and and medical medical communities. communities. EGL EGL Genetics Genetics is is one one of of only only a afew few clinical clinical diagnostic diagnostic laboratories laboratories to to openly openly share share data data withwith the the NCBI NCBI freely freely available available public public database database ClinVar ClinVar (>35,000 (>35,000 variants variants on on >1700 >1700 genes) genes) and and is isalso also the the only only laboratory laboratory with with a a frefree oen olinnlein dea dtabtaabsaes (eE m(EVmCVlaCslas)s,s f)e, afetuatruinrgin ag vaa vraiarniatn ctl acslasisfiscifiactiaotino sne saercahrc ahn adn rde rpeoprot rrte rqeuqeuset sint tinetrefarcfaec, ew, hwichhic fha cfailcitialiteatse rsa praidp id interactiveinteractive curation curation and and reporting reporting of of variants. -

COVID-19—The Potential Beneficial Therapeutic Effects of Spironolactone During SARS-Cov-2 Infection
pharmaceuticals Review COVID-19—The Potential Beneficial Therapeutic Effects of Spironolactone during SARS-CoV-2 Infection Katarzyna Kotfis 1,* , Kacper Lechowicz 1 , Sylwester Drozd˙ zal˙ 2 , Paulina Nied´zwiedzka-Rystwej 3 , Tomasz K. Wojdacz 4, Ewelina Grywalska 5 , Jowita Biernawska 6, Magda Wi´sniewska 7 and Miłosz Parczewski 8 1 Department of Anesthesiology, Intensive Therapy and Acute Intoxications, Pomeranian Medical University in Szczecin, 70-111 Szczecin, Poland; [email protected] 2 Department of Pharmacokinetics and Monitored Therapy, Pomeranian Medical University, 70-111 Szczecin, Poland; [email protected] 3 Institute of Biology, University of Szczecin, 71-412 Szczecin, Poland; [email protected] 4 Independent Clinical Epigenetics Laboratory, Pomeranian Medical University, 71-252 Szczecin, Poland; [email protected] 5 Department of Clinical Immunology and Immunotherapy, Medical University of Lublin, 20-093 Lublin, Poland; [email protected] 6 Department of Anesthesiology and Intensive Therapy, Pomeranian Medical University in Szczecin, 71-252 Szczecin, Poland; [email protected] 7 Clinical Department of Nephrology, Transplantology and Internal Medicine, Pomeranian Medical University, 70-111 Szczecin, Poland; [email protected] 8 Department of Infectious, Tropical Diseases and Immune Deficiency, Pomeranian Medical University in Szczecin, 71-455 Szczecin, Poland; [email protected] * Correspondence: katarzyna.kotfi[email protected]; Tel.: +48-91-466-11-44 Abstract: In March 2020, coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 was declared Citation: Kotfis, K.; Lechowicz, K.; a global pandemic by the World Health Organization (WHO). The clinical course of the disease is Drozd˙ zal,˙ S.; Nied´zwiedzka-Rystwej, unpredictable but may lead to severe acute respiratory infection (SARI) and pneumonia leading to P.; Wojdacz, T.K.; Grywalska, E.; acute respiratory distress syndrome (ARDS). -

Ras Gtpase Chemi ELISA Kit Catalog No
Ras GTPase Chemi ELISA Kit Catalog No. 52097 (Version B3) Active Motif North America 1914 Palomar Oaks Way, Suite 150 Carlsbad, California 92008, USA Toll free: 877 222 9543 Telephone: 760 431 1263 Fax: 760 431 1351 Active Motif Europe Waterloo Atrium Drève Richelle 167 – boîte 4 BE-1410 Waterloo, Belgium UK Free Phone: 0800 169 31 47 France Free Phone: 0800 90 99 79 Germany Free Phone: 0800 181 99 10 Telephone: +32 (0)2 653 0001 Fax: +32 (0)2 653 0050 Active Motif Japan Azuma Bldg, 7th Floor 2-21 Ageba-Cho, Shinjuku-Ku Tokyo, 162-0824, Japan Telephone: +81 3 5225 3638 Fax: +81 3 5261 8733 Active Motif China 787 Kangqiao Road Building 10, Suite 202, Pudong District Shanghai, 201315, China Telephone: (86)-21-20926090 Hotline: 400-018-8123 Copyright 2021 Active Motif, Inc. www.activemotif.com Information in this manual is subject to change without notice and does not constitute a commit- ment on the part of Active Motif, Inc. It is supplied on an “as is” basis without any warranty of any kind, either explicit or implied. Information may be changed or updated in this manual at any time. This documentation may not be copied, transferred, reproduced, disclosed, or duplicated, in whole or in part, without the prior written consent of Active Motif, Inc. This documentation is proprietary information and protected by the copyright laws of the United States and interna- tional treaties. The manufacturer of this documentation is Active Motif, Inc. © 2021 Active Motif, Inc., 1914 Palomar Oaks Way, Suite 150; Carlsbad, CA 92008. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Identification of Synaptic Proteins and Their Isoform Mrnas In
Proc. Natl. Acad. Sci. USA Vol. 91, pp. 12487-12491, December 1994 Cell Biology Identification of synaptic proteins and their isoform mRNAs in compartments of pancreatic endocrine cells (exocytosis/secretion/insulin/diabetes) GUNILLA JACOBSSON*, ANDREW J. BEANt, RICHARD H. SCHELLERt, LISA JUNTTI-BERGGRENt, JUDE T. DEENEYt, PER-OLOF BERGGRENt AND BJORN MEISTER*§ *Department of Neuroscience and tRolf Luft's Center for Diabetes Research, Department of Molecular Medicine, Karolinska Institute, S-171 77 Stockholm, Sweden; and tDepartment of Molecular and Cellular Physiology, Howard Hughes Medical Institute, Beckman Center, Stanford University, Stanford, CA 94305 Communicated by Tomas Hokfelt, August 30, 1994 ABSTRACT Several proteins that are of importance for clostridial neurotoxins, including tetanus toxin and botuli- membrane trafficking in the nerve terminal have recently been num neurotoxin B, whereas botulinum neurotoxins D and F characterized. We have used Western blot and immunohis- are capable of cleaving both forms of VAMP (10-12). tochemistry to show that synaptotagmin, synaptobrevin/VAMP VAMP-1 and VAMP-2 are encoded by two distinct genes (13) (vesicle-associated membrane protein), SNAP-25 (synaptosom- and are differentially expressed in the nervous system (14). al-associated protein of 25 kDa), and syntaxin proteins are Cellubrevin is a homologue of VAMP, which is present in a present in cells of the islets of Langerhans in the endocrine wide variety of tissues and may be a membrane trafficking pancreas. Synaptotagmin-like immunoreactivity (-LI) was lo- protein of a constitutively recycling pathway (15). calized to granules within the cytoplasm of a few endocrine cells In contrast to synaptotagmin and VAMP, the synaptoso- located in the periphery of the islets, identified as somatostatin- mal-associated protein of 25 kDa (SNAP-25) is located at the containing cells, and in many nerve fibers within the islets. -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Hras Intracellular Trafficking and Signal Transduction Jodi Ho-Jung Mckay Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 HRas intracellular trafficking and signal transduction Jodi Ho-Jung McKay Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cancer Biology Commons, Cell Biology Commons, Genetics and Genomics Commons, and the Medical Cell Biology Commons Recommended Citation McKay, Jodi Ho-Jung, "HRas intracellular trafficking and signal transduction" (2007). Retrospective Theses and Dissertations. 13946. https://lib.dr.iastate.edu/rtd/13946 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. HRas intracellular trafficking and signal transduction by Jodi Ho-Jung McKay A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics Program of Study Committee: Janice E. Buss, Co-major Professor Linda Ambrosio, Co-major Professor Diane Bassham Drena Dobbs Ted Huiatt Iowa State University Ames, Iowa 2007 Copyright © Jodi Ho-Jung McKay, 2007. All rights reserved. UMI Number: 3274881 Copyright 2007 by McKay, Jodi Ho-Jung All rights reserved. UMI Microform 3274881 Copyright 2008 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. -

Variable Priming of a Docked Synaptic Vesicle PNAS PLUS
Variable priming of a docked synaptic vesicle PNAS PLUS Jae Hoon Junga,b,c, Joseph A. Szulea,c, Robert M. Marshalla,c, and Uel J. McMahana,c,1 aDepartment of Neurobiology, Stanford University School of Medicine, Stanford, CA 94305; bDepartment of Physics, Stanford University School of Humanities and Sciences, Stanford, CA 94305; and cDepartment of Biology, Texas A&M University, College Station, TX 77845 Edited by Thomas S. Reese, National Institutes of Health, Bethesda, MD, and approved January 12, 2016 (received for review November 30, 2015) The priming of a docked synaptic vesicle determines the proba- transition. Biochemical and electrophysiological approaches have bility of its membrane (VM) fusing with the presynaptic membrane provided evidence that priming is mediated by interactions be- (PM) when a nerve impulse arrives. To gain insight into the nature tween the SNARE proteins and their regulators (7, 12–14, 24) and of priming, we searched by electron tomography for structural can involve differences in positioning of docked SVs relative to + relationships correlated with fusion probability at active zones of Ca2 channels (25). Biochemistry has also led to the suggestion axon terminals at frog neuromuscular junctions. For terminals that primed SVs may become deprimed (26). fixed at rest, the contact area between the VM of docked vesicles We have previously shown by electron tomography on frog and PM varied >10-fold with a normal distribution. There was no neuromuscular junctions (NMJs) fixed at rest that there are, for merging of the membranes. For terminals fixed during repetitive docked SVs, variations in the extent of the VM–PM contact area evoked synaptic transmission, the normal distribution of contact and in the length of the several AZM macromolecules linking areas was shifted to the left, due in part to a decreased number of the VM to the PM, the so-called ribs, pegs, and pins (2, 27). -

Ciliopathies Gene Panel
Ciliopathies Gene Panel Contact details Introduction Regional Genetics Service The ciliopathies are a heterogeneous group of conditions with considerable phenotypic overlap. Levels 4-6, Barclay House These inherited diseases are caused by defects in cilia; hair-like projections present on most 37 Queen Square cells, with roles in key human developmental processes via their motility and signalling functions. Ciliopathies are often lethal and multiple organ systems are affected. Ciliopathies are London, WC1N 3BH united in being genetically heterogeneous conditions and the different subtypes can share T +44 (0) 20 7762 6888 many clinical features, predominantly cystic kidney disease, but also retinal, respiratory, F +44 (0) 20 7813 8578 skeletal, hepatic and neurological defects in addition to metabolic defects, laterality defects and polydactyly. Their clinical variability can make ciliopathies hard to recognise, reflecting the ubiquity of cilia. Gene panels currently offer the best solution to tackling analysis of genetically Samples required heterogeneous conditions such as the ciliopathies. Ciliopathies affect approximately 1:2,000 5ml venous blood in plastic EDTA births. bottles (>1ml from neonates) Ciliopathies are generally inherited in an autosomal recessive manner, with some autosomal Prenatal testing must be arranged dominant and X-linked exceptions. in advance, through a Clinical Genetics department if possible. Referrals Amniotic fluid or CV samples Patients presenting with a ciliopathy; due to the phenotypic variability this could be a diverse set should be sent to Cytogenetics for of features. For guidance contact the laboratory or Dr Hannah Mitchison dissecting and culturing, with ([email protected]) / Prof Phil Beales ([email protected]) instructions to forward the sample to the Regional Molecular Genetics Referrals will be accepted from clinical geneticists and consultants in nephrology, metabolic, laboratory for analysis respiratory and retinal diseases. -
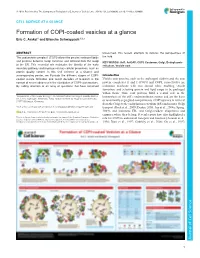
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Deciphering the Nature of Trp73 Isoforms in Mouse
cancers Article Deciphering the Nature of Trp73 Isoforms in Mouse Embryonic Stem Cell Models: Generation of Isoform-Specific Deficient Cell Lines Using the CRISPR/Cas9 Gene Editing System Lorena López-Ferreras 1,2,†, Nicole Martínez-García 1,3,†, Laura Maeso-Alonso 1,2,‡, Marta Martín-López 1,4,‡, Ángela Díez-Matilla 1,‡ , Javier Villoch-Fernandez 1,2, Hugo Alonso-Olivares 1,2, Margarita M. Marques 3,5,* and Maria C. Marin 1,2,* 1 Instituto de Biomedicina (IBIOMED), Universidad de León, 24071 León, Spain; [email protected] (L.L.-F.); [email protected] (N.M.-G.); [email protected] (L.M.-A.); [email protected] (M.M.-L.); [email protected] (Á.D.-M.); [email protected] (J.V.-F.); [email protected] (H.A.-O.) 2 Departamento de Biología Molecular, Universidad de León, 24071 León, Spain 3 Departamento de Producción Animal, Universidad de León, 24071 León, Spain 4 Biomar Microbial Technologies, Parque Tecnológico de León, Armunia, 24009 León, Spain 5 Instituto de Desarrollo Ganadero y Sanidad Animal (INDEGSAL), Universidad de León, 24071 León, Spain * Correspondence: [email protected] (M.M.M.); [email protected] (M.C.M.); Tel.: +34-987-291757 Citation: López-Ferreras, L.; (M.M.M.); +34-987-291490 (M.C.M.) Martínez-García, N.; Maeso-Alonso, † Equal contribution. ‡ Equal contribution. L.; Martín-López, M.; Díez-Matilla, Á.; Villoch-Fernandez, J.; Simple Summary: The Trp73 gene is involved in the regulation of multiple biological processes Alonso-Olivares, H.; Marques, M.M.; Marin, M.C. Deciphering the Nature such as response to stress, differentiation and tissue architecture.