Identification of Viruses Associated with Larvae of the Dragonfly
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Viral Infection in the Copepods Labidocera Aestiva And
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2012 Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida Darren Stephenson Dunlap University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Other Oceanography and Atmospheric Sciences and Meteorology Commons, and the Virology Commons Scholar Commons Citation Dunlap, Darren Stephenson, "Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida" (2012). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/4032 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Evidence of Viruses in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida By Darren S. Dunlap A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science College of Marine Science University of South Florida Major Professor: Mya Breitbart, Ph.D Kendra Daly, Ph.D Ian Hewson, Ph.D Date of Approval: March 19, 2012 Key Words: Copepods, Single-stranded DNA Viruses, Mesozooplankton, Transmission Electron Microscopy, Metagenomics Copyright © 2012, Darren Stephenson Dunlap DEDICATION None of this would have been possible without the generous love and support of my entire family over the years. My parents, Steve and Jill Dunlap, have always encouraged my pursuits with support and love, and their persistence of throwing me into lakes and rivers is largely responsible for my passion for Marine Science. -

Identification of Capsid/Coat Related Protein Folds and Their Utility for Virus Classification
ORIGINAL RESEARCH published: 10 March 2017 doi: 10.3389/fmicb.2017.00380 Identification of Capsid/Coat Related Protein Folds and Their Utility for Virus Classification Arshan Nasir 1, 2 and Gustavo Caetano-Anollés 1* 1 Department of Crop Sciences, Evolutionary Bioinformatics Laboratory, University of Illinois at Urbana-Champaign, Urbana, IL, USA, 2 Department of Biosciences, COMSATS Institute of Information Technology, Islamabad, Pakistan The viral supergroup includes the entire collection of known and unknown viruses that roam our planet and infect life forms. The supergroup is remarkably diverse both in its genetics and morphology and has historically remained difficult to study and classify. The accumulation of protein structure data in the past few years now provides an excellent opportunity to re-examine the classification and evolution of viruses. Here we scan completely sequenced viral proteomes from all genome types and identify protein folds involved in the formation of viral capsids and virion architectures. Viruses encoding similar capsid/coat related folds were pooled into lineages, after benchmarking against published literature. Remarkably, the in silico exercise reproduced all previously described members of known structure-based viral lineages, along with several proposals for new Edited by: additions, suggesting it could be a useful supplement to experimental approaches and Ricardo Flores, to aid qualitative assessment of viral diversity in metagenome samples. Polytechnic University of Valencia, Spain Keywords: capsid, virion, protein structure, virus taxonomy, SCOP, fold superfamily Reviewed by: Mario A. Fares, Consejo Superior de Investigaciones INTRODUCTION Científicas(CSIC), Spain Janne J. Ravantti, The last few years have dramatically increased our knowledge about viral systematics and University of Helsinki, Finland evolution. -

And Giant Guitarfish (Rhynchobatus Djiddensis)
VIRAL DISCOVERY IN BLUEGILL SUNFISH (LEPOMIS MACROCHIRUS) AND GIANT GUITARFISH (RHYNCHOBATUS DJIDDENSIS) BY HISTOPATHOLOGY EVALUATION, METAGENOMIC ANALYSIS AND NEXT GENERATION SEQUENCING by JENNIFER ANNE DILL (Under the Direction of Alvin Camus) ABSTRACT The rapid growth of aquaculture production and international trade in live fish has led to the emergence of many new diseases. The introduction of novel disease agents can result in significant economic losses, as well as threats to vulnerable wild fish populations. Losses are often exacerbated by a lack of agent identification, delay in the development of diagnostic tools and poor knowledge of host range and susceptibility. Examples in bluegill sunfish (Lepomis macrochirus) and the giant guitarfish (Rhynchobatus djiddensis) will be discussed here. Bluegill are popular freshwater game fish, native to eastern North America, living in shallow lakes, ponds, and slow moving waterways. Bluegill experiencing epizootics of proliferative lip and skin lesions, characterized by epidermal hyperplasia, papillomas, and rarely squamous cell carcinoma, were investigated in two isolated poopulations. Next generation genomic sequencing revealed partial DNA sequences of an endogenous retrovirus and the entire circular genome of a novel hepadnavirus. Giant Guitarfish, a rajiform elasmobranch listed as ‘vulnerable’ on the IUCN Red List, are found in the tropical Western Indian Ocean. Proliferative skin lesions were observed on the ventrum and caudal fin of a juvenile male quarantined at a public aquarium following international shipment. Histologically, lesions consisted of papillomatous epidermal hyperplasia with myriad large, amphophilic, intranuclear inclusions. Deep sequencing and metagenomic analysis produced the complete genomes of two novel DNA viruses, a typical polyomavirus and a second unclassified virus with a 20 kb genome tentatively named Colossomavirus. -

Data Mining Cdnas Reveals Three New Single Stranded RNA Viruses in Nasonia (Hymenoptera: Pteromalidae)
Insect Molecular Biology Insect Molecular Biology (2010), 19 (Suppl. 1), 99–107 doi: 10.1111/j.1365-2583.2009.00934.x Data mining cDNAs reveals three new single stranded RNA viruses in Nasonia (Hymenoptera: Pteromalidae) D. C. S. G. Oliveira*, W. B. Hunter†, J. Ng*, Small viruses with a positive-sense single-stranded C. A. Desjardins*, P. M. Dang‡ and J. H. Werren* RNA (ssRNA) genome, and no DNA stage, are known as *Department of Biology, University of Rochester, picornaviruses (infecting vertebrates) or picorna-like Rochester, NY, USA; †United States Department of viruses (infecting non-vertebrates). Recently, the order Agriculture, Agricultural Research Service, US Picornavirales was formally characterized to include most, Horticultural Research Laboratory, Fort Pierce, FL, USA; but not all, ssRNA viruses (Le Gall et al., 2008). Among and ‡United States Department of Agriculture, other typical characteristics – e.g. a small icosahedral Agricultural Research Service, NPRU, Dawson, GA, capsid with a pseudo-T = 3 symmetry and a 7–12 kb USA genome made of one or two RNA segments – the Picor- navirales genome encodes a polyprotein with a replication Abstractimb_934 99..108 module that includes a helicase, a protease, and an RNA- dependent RNA polymerase (RdRp), in this order (see Le We report three novel small RNA viruses uncovered Gall et al., 2008 for details). Pathogenicity of the infections from cDNA libraries from parasitoid wasps in the can vary broadly from devastating epidemics to appar- genus Nasonia. The genome of this kind of virus ently persistent commensal infections. Several human Ј is a positive-sense single-stranded RNA with a 3 diseases, from hepatitis A to the common cold (e.g. -

On Fenn's and Whixall Moss, Shropshire, UK
Recapture rates and habitat associations of White-faced Darter Leucorhinnia dubia on Fenn's and Whixall Moss, Shropshire, UK Item Type Article Authors Davies, Rachel; von Hardenberg, Achaz; Geary, Matthew Citation Recapture rates and habitat associations of White-faced Darter Leucorhinnia dubia on Fenn's and Whixall Moss, Shropshire, UK R Davies, A von Hardenberg, M Geary Journal of the British Dragonfly Society 34 (2), 89-101 Publisher British Dragonfly Society Journal Journal of the British Dragonfly Society Rights Attribution 3.0 United States Download date 28/09/2021 18:54:03 Item License https://creativecommons.org/licenses/by-nc-nd/4.0/ Link to Item http://hdl.handle.net/10034/621511 Recapture rates and habitat associations of Leucorrhinia dubia (White-faced Darter; Vander Linden) on Fenn’s and Whixall Moss, Shropshire, UK Rachel Davies, Achaz von Hardenberg & Matthew Geary* Conservation Biology Research Group, Department of Biological Science, University of Chester, Chester, CH1 4BJ *Corresponding author: [email protected] Abstract Land-use change and habitat loss are important drivers of biodiversity decline at both global and local scales. To protect species from the impacts of land-use change it is important to understand the population dynamics and habitat associations across these scales. Here we present an investigation into the survival and habitat preferences of Leucorrhinia dubia at the local scale at Fenn’s and Whixall Moss, Shropshire, UK. We used mark-release-recapture methods to investigate survival and used sightings of individual dragonflies along with habitat data to investigate habitat preference. We found that survival between capture- visits was very low and that L. -

Frequent Occurrence of Mungbean Yellow Mosaic India Virus in Tomato Leaf Curl Disease Afected Tomato in Oman M
www.nature.com/scientificreports OPEN Frequent occurrence of Mungbean yellow mosaic India virus in tomato leaf curl disease afected tomato in Oman M. S. Shahid 1*, M. Shafq 1, M. Ilyas2, A. Raza1, M. N. Al-Sadrani1, A. M. Al-Sadi 1 & R. W. Briddon 3 Next generation sequencing (NGS) of DNAs amplifed by rolling circle amplifcation from 6 tomato (Solanum lycopersicum) plants with leaf curl symptoms identifed a number of monopartite begomoviruses, including Tomato yellow leaf curl virus (TYLCV), and a betasatellite (Tomato leaf curl betasatellite [ToLCB]). Both TYLCV and ToLCB have previously been identifed infecting tomato in Oman. Surprisingly the NGS results also suggested the presence of the bipartite, legume-adapted begomovirus Mungbean yellow mosaic Indian virus (MYMIV). The presence of MYMIV was confrmed by cloning and Sanger sequencing from four of the six plants. A wider analysis by PCR showed MYMIV infection of tomato in Oman to be widespread. Inoculation of plants with full-length clones showed the host range of MYMIV not to extend to Nicotiana benthamiana or tomato. Inoculation to N. benthamiana showed TYLCV to be capable of maintaining MYMIV in both the presence and absence of the betasatellite. In tomato MYMIV was only maintained by TYLCV in the presence of the betasatellite and then only at low titre and efciency. This is the frst identifcation of TYLCV with ToLCB and the legume adapted bipartite begomovirus MYMIV co-infecting tomato. This fnding has far reaching implications. TYLCV has spread around the World from its origins in the Mediterranean/Middle East, in some instances, in live tomato planting material. -
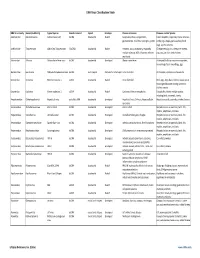
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Viruses in Transplantation - Not Always Enemies
Viruses in transplantation - not always enemies Virome and transplantation ECCMID 2018 - Madrid Prof. Laurent Kaiser Head Division of Infectious Diseases Laboratory of Virology Geneva Center for Emerging Viral Diseases University Hospital of Geneva ESCMID eLibrary © by author Conflict of interest None ESCMID eLibrary © by author The human virome: definition? Repertoire of viruses found on the surface of/inside any body fluid/tissue • Eukaryotic DNA and RNA viruses • Prokaryotic DNA and RNA viruses (phages) 25 • The “main” viral community (up to 10 bacteriophages in humans) Haynes M. 2011, Metagenomic of the human body • Endogenous viral elements integrated into host chromosomes (8% of the human genome) • NGS is shaping the definition Rascovan N et al. Annu Rev Microbiol 2016;70:125-41 Popgeorgiev N et al. Intervirology 2013;56:395-412 Norman JM et al. Cell 2015;160:447-60 ESCMID eLibraryFoxman EF et al. Nat Rev Microbiol 2011;9:254-64 © by author Viruses routinely known to cause diseases (non exhaustive) Upper resp./oropharyngeal HSV 1 Influenza CNS Mumps virus Rhinovirus JC virus RSV Eye Herpes viruses Parainfluenza HSV Measles Coronavirus Adenovirus LCM virus Cytomegalovirus Flaviviruses Rabies HHV6 Poliovirus Heart Lower respiratory HTLV-1 Coxsackie B virus Rhinoviruses Parainfluenza virus HIV Coronaviruses Respiratory syncytial virus Parainfluenza virus Adenovirus Respiratory syncytial virus Coronaviruses Gastro-intestinal Influenza virus type A and B Human Bocavirus 1 Adenovirus Hepatitis virus type A, B, C, D, E Those that cause -

Identification of a Nanovirus-Alphasatellite Complex in Sophora Alopecuroides
Accepted Manuscript Title: Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides Author: Jahangir Heydarnejad Mehdi Kamali Hossain Massumi Anders Kvarnheden Maketalena F. Male Simona Kraberger Daisy Stainton Darren P. Martin Arvind Varsani PII: S0168-1702(17)30009-6 DOI: http://dx.doi.org/doi:10.1016/j.virusres.2017.03.023 Reference: VIRUS 97113 To appear in: Virus Research Received date: 5-1-2017 Revised date: 15-3-2017 Accepted date: 18-3-2017 Please cite this article as: Heydarnejad, J., Kamali, M., Massumi, H., Kvarnheden, A., Male, M.F., Kraberger, S., Stainton, D., Martin, D.P., Varsani, A.,Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides, Virus Research (2017), http://dx.doi.org/10.1016/j.virusres.2017.03.023 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides 2 Jahangir Heydarnejad 1* , Mehdi Kamali 1, Hossain Massumi 1, Anders Kvarnheden 2, Maketalena 3 F. Male 3, Simona Kraberger 3,4 , Daisy Stainton 3,5 , Darren P. Martin 6 and Arvind Varsani 3,7,8* 4 5 1Department of Plant Protection, College -

The Complete Genome of an Endogenous Nimavirus (Nimav-1 Lva) from the Pacific Whiteleg Shrimp Penaeus (Litopenaeus) Vannamei
G C A T T A C G G C A T genes Article The Complete Genome of an Endogenous Nimavirus (Nimav-1_LVa) From the Pacific Whiteleg Shrimp Penaeus (Litopenaeus) Vannamei Weidong Bao 1,* , Kathy F. J. Tang 2 and Acacia Alcivar-Warren 3,4,* 1 Genetic Information Research Institute, 20380 Town Center Lane, Suite 240, Cupertino, CA 95014, USA 2 Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, 106 Nanjing Road, Qingdao 266071, China; [email protected] 3 Fundación para la Conservation de la Biodiversidad Acuática y Terrestre (FUCOBI), Quito EC1701, Ecuador 4 Environmental Genomics Inc., ONE HEALTH Epigenomics Educational Initiative, P.O. Box 196, Southborough, MA 01772, USA * Correspondence: [email protected] (W.B.); [email protected] (A.A.-W.) Received: 17 December 2019; Accepted: 9 January 2020; Published: 14 January 2020 Abstract: White spot syndrome virus (WSSV), the lone virus of the genus Whispovirus under the family Nimaviridae, is one of the most devastating viruses affecting the shrimp farming industry. Knowledge about this virus, in particular, its evolution history, has been limited, partly due to its large genome and the lack of other closely related free-living viruses for comparative studies. In this study, we reconstructed a full-length endogenous nimavirus consensus genome, Nimav-1_LVa (279,905 bp), in the genome sequence of Penaeus (Litopenaeus) vannamei breed Kehai No. 1 (ASM378908v1). This endogenous virus seemed to insert exclusively into the telomeric pentanucleotide microsatellite (TAACC/GGTTA)n. It encoded 117 putative genes, with some containing introns, such as g012 (inhibitor of apoptosis, IAP), g046 (crustacean hyperglycemic hormone, CHH), g155 (innexin), g158 (Bax inhibitor 1 like). -

Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics
viruses Article Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics Ashleigh F. Porter 1, Mang Shi 1, John-Sebastian Eden 1,2 , Yong-Zhen Zhang 3,4 and Edward C. Holmes 1,3,* 1 Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life & Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia; [email protected] (A.F.P.); [email protected] (M.S.); [email protected] (J.-S.E.) 2 Centre for Virus Research, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia 3 Shanghai Public Health Clinical Center and School of Public Health, Fudan University, Shanghai 201500, China; [email protected] 4 Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China * Correspondence: [email protected]; Tel.: +61-2-9351-5591 Received: 17 October 2019; Accepted: 23 November 2019; Published: 25 November 2019 Abstract: DNA viruses comprise a wide array of genome structures and infect diverse host species. To date, most studies of DNA viruses have focused on those with the strongest disease associations. Accordingly, there has been a marked lack of sampling of DNA viruses from invertebrates. Bulk RNA sequencing has resulted in the discovery of a myriad of novel RNA viruses, and herein we used this methodology to identify actively transcribing DNA viruses in meta-transcriptomic libraries of diverse invertebrate species. Our analysis revealed high levels of phylogenetic diversity in DNA viruses, including 13 species from the Parvoviridae, Circoviridae, and Genomoviridae families of single-stranded DNA virus families, and six double-stranded DNA virus species from the Nudiviridae, Polyomaviridae, and Herpesviridae, for which few invertebrate viruses have been identified to date. -

Diversity of Large DNA Viruses of Invertebrates ⇑ Trevor Williams A, Max Bergoin B, Monique M
Journal of Invertebrate Pathology 147 (2017) 4–22 Contents lists available at ScienceDirect Journal of Invertebrate Pathology journal homepage: www.elsevier.com/locate/jip Diversity of large DNA viruses of invertebrates ⇑ Trevor Williams a, Max Bergoin b, Monique M. van Oers c, a Instituto de Ecología AC, Xalapa, Veracruz 91070, Mexico b Laboratoire de Pathologie Comparée, Faculté des Sciences, Université Montpellier, Place Eugène Bataillon, 34095 Montpellier, France c Laboratory of Virology, Wageningen University, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands article info abstract Article history: In this review we provide an overview of the diversity of large DNA viruses known to be pathogenic for Received 22 June 2016 invertebrates. We present their taxonomical classification and describe the evolutionary relationships Revised 3 August 2016 among various groups of invertebrate-infecting viruses. We also indicate the relationships of the Accepted 4 August 2016 invertebrate viruses to viruses infecting mammals or other vertebrates. The shared characteristics of Available online 31 August 2016 the viruses within the various families are described, including the structure of the virus particle, genome properties, and gene expression strategies. Finally, we explain the transmission and mode of infection of Keywords: the most important viruses in these families and indicate, which orders of invertebrates are susceptible to Entomopoxvirus these pathogens. Iridovirus Ó Ascovirus 2016 Elsevier Inc. All rights reserved. Nudivirus Hytrosavirus Filamentous viruses of hymenopterans Mollusk-infecting herpesviruses 1. Introduction in the cytoplasm. This group comprises viruses in the families Poxviridae (subfamily Entomopoxvirinae) and Iridoviridae. The Invertebrate DNA viruses span several virus families, some of viruses in the family Ascoviridae are also discussed as part of which also include members that infect vertebrates, whereas other this group as their replication starts in the nucleus, which families are restricted to invertebrates.