Design Automation in Synthetic Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Informa Tics for Genome Analysis, Biomarkers,And Ta Rget Disco Very
Register by March 14 and Save up to $200! Final Agenda Cambridge Healthtech Institute’s Sixth Annual April 28 – 30, 2008 • World Trade Center • Boston, MA Keynote Presentations by: Concurrent Tracks: • Informatics and IT Infrastructure & Operations • Informatics for Genome Analysis, Biomarkers, and Target Discovery Linda Avey co-Founder • Predictive and in silico Science 23andMe, Inc. • eChemistry Solutions • Clinical and Medical Informatics Joshua Boger, Ph.D. President & Chief Executive Offi cer Keynote Panel: A Must-Attend Event for the Life Vertex Pharmaceuticals, Inc. The Future of Personal Genomics: A special plenary panel discussion featuring George Church,Science Ph.D., Harvard IT Medical& Informatics School Community Dietrich Stephan, Ph.D., Navigenics John Reynders, Ph.D. Jeffrey M. Drazen, M.D., New England Journal of Medicine/Harvard Vice President & Chief Information Offi cer Medical School Johnson & Johnson, Pharma R&D Fred D. Ledley, M.D., Bentley College ...and more 2008 Benjamin Franklin Award EEventvent FFeatures:eatures: presented by Bioinformatics.org • Access All Five Tracks for One Price • Network with 1,500+ Delegates Platinum Sponsors: • Hear 85+ Technology and Scientifi c Presentations • Choose from Four Pre-conference Workshops, the Oracle Life Sciences and Healthcare User Group Meeting, and Symyx Software Symposium 2008 Gold Sponsors: • Attend Bio-IT World’s Best Practices Awards NEW Bronze Sponsor: • Connect with Attendees Using CHI’s Intro-Net Corporate Support: • Participate in the Poster Competition • See the Winners of the following 2008 Awards: Offi cial Publication: Benjamin Franklin Best of Show Lead Sponsoring Publications: Best Practices • View Who’s Who & What’s New in the Exhibit Hall • And Much More! www.Bio-ITWorldExpo.com Organized & Managed by: Cambridge Healthtech Institute 250 First Avenue, Suite 300, Needham, MA 02494 • Phone: 781-972-5400 • Fax: 781-972-5425 • Toll-free in the U.S. -

An Open-Sourced Bioinformatic Pipeline for the Processing of Next-Generation Sequencing Derived Nucleotide Reads
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.20.050369; this version posted May 28, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. An open-sourced bioinformatic pipeline for the processing of Next-Generation Sequencing derived nucleotide reads: Identification and authentication of ancient metagenomic DNA Thomas C. Collin1, *, Konstantina Drosou2, 3, Jeremiah Daniel O’Riordan4, Tengiz Meshveliani5, Ron Pinhasi6, and Robin N. M. Feeney1 1School of Medicine, University College Dublin, Ireland 2Division of Cell Matrix Biology Regenerative Medicine, University of Manchester, United Kingdom 3Manchester Institute of Biotechnology, School of Earth and Environmental Sciences, University of Manchester, United Kingdom [email protected] 5Institute of Paleobiology and Paleoanthropology, National Museum of Georgia, Tbilisi, Georgia 6Department of Evolutionary Anthropology, University of Vienna, Austria *Corresponding Author Abstract The emerging field of ancient metagenomics adds to these Bioinformatic pipelines optimised for the processing and as- processing complexities with the need for additional steps sessment of metagenomic ancient DNA (aDNA) are needed in the separation and authentication of ancient sequences from modern sequences. Currently, there are few pipelines for studies that do not make use of high yielding DNA cap- available for the analysis of ancient metagenomic DNA ture techniques. These bioinformatic pipelines are tradition- 1 4 ally optimised for broad aDNA purposes, are contingent on (aDNA) ≠ The limited number of bioinformatic pipelines selection biases and are associated with high costs. -

Research Laboratory of Electronics
Research Laboratory of Electronics The Research Laboratory of Electronics (RLE) at MIT is a vibrant intellectual community and was one of the Institute’s earliest modern interdepartmental academic research centers. RLE research encompasses both basic and applied science and engineering in an extensive range of natural and man-made phenomena. Integral to RLE’s efforts is the furthering of scientific understanding and leading innovation to provide great service to society. The lab’s research spans the fundamentals of quantum physics and information theory to synthetic biology and power electronics and extends to novel engineering applications, including those that produce significant advances in communication systems or enable remote sensing from aircraft and spacecraft, and the development of new biomaterials and innovations in diagnostics and treatment of human diseases. RLE was founded in 1946 following the groundbreaking research that led to the development of ultra-high-frequency radar, a technology that changed the course of World War II. It was home to some of the great discoveries made in the 20th century at MIT. Cognizant of its rich history and focus on maintaining its position as MIT’s leading interdisciplinary research organization, RLE fosters a stimulating and supportive environment for innovative research and impact. What distinguishes RLE today from all other entities at MIT is its breadth of intellectual pursuit and diversity of research themes. The lab supports its researchers by providing a wide array of services spanning financial and human resources (HR), information technology, and renovations. It is fiscally independent and maintains a high level of loyalty from its principal investigators (PIs) and staff. -

View Publication
Downloaded from rsif.royalsocietypublishing.org on 20 April 2009 Towards programming languages for genetic engineering of living cells Michael Pedersen and Andrew Phillips J. R. Soc. Interface published online 15 April 2009 doi: 10.1098/rsif.2008.0516.focus Supplementary data "Data Supplement" http://rsif.royalsocietypublishing.org/content/suppl/2009/04/15/rsif.2008.0516.focus.D C1.html References This article cites 19 articles, 7 of which can be accessed free http://rsif.royalsocietypublishing.org/content/early/2009/04/14/rsif.2008.0516.focus.full. html#ref-list-1 P<P Published online 15 April 2009 in advance of the print journal. Subject collections Articles on similar topics can be found in the following collections synthetic biology (9 articles) Email alerting service Receive free email alerts when new articles cite this article - sign up in the box at the top right-hand corner of the article or click here Advance online articles have been peer reviewed and accepted for publication but have not yet appeared in the paper journal (edited, typeset versions may be posted when available prior to final publication). Advance online articles are citable and establish publication priority; they are indexed by PubMed from initial publication. Citations to Advance online articles must include the digital object identifier (DOIs) and date of initial publication. To subscribe to J. R. Soc. Interface go to: http://rsif.royalsocietypublishing.org/subscriptions This journal is © 2009 The Royal Society Downloaded from rsif.royalsocietypublishing.org on 20 April 2009 J. R. Soc. Interface doi:10.1098/rsif.2008.0516.focus Published online Towards programming languages for genetic engineering of living cells Michael Pedersen1,2 and Andrew Phillips1,* 1Microsoft Research Cambridge, Cambridge CB3 0FB, UK 2LFCS, School of Informatics, University of Edinburgh, Edinburgh EH8 9AB, UK Synthetic biology aims at producing novel biological systems to carry out some desired and well-defined functions. -

SYNTHETIC BIOLOGY and ITS POTENTIAL IMPLICATIONS for BIOTRADE and ACCESS and BENEFIT-SHARING ©2019, United Nations Conference on Trade and Development
UNITED NATIONS CONFERENCE ON TRADE AND DEVELOPMENT SYNTHETIC BIOLOGY AND ITS POTENTIAL IMPLICATIONS FOR BIOTRADE AND ACCESS AND BENEFIT-SHARING ©2019, United Nations Conference on Trade and Development. All rights reserved. The trends, figures and views expressed in this publication are those of UNCTAD and do not necessarily represent the views of its member States. The designations employed and the presentation of material on any map in this work do not imply the expression of any opinion whatsoever on the part of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. This study can be freely cited provided appropriate acknowledgment is given to UNCTAD. For further information on UNCTAD’s BioTrade Initiative please consult the following website: http://www.unctad. org/biotrade or contact us at: [email protected] This publication has not been formally edited. UNCTAD/DITC/TED/INF/2019/12 AND ITS POTENTIAL IMPLICATIONS FOR BIOTRADE AND ACCESS AND BENEFIT-SHARING iii Contents Acknowledgements ................................................................................................................................iv Abbreviations ...........................................................................................................................................v EXECUTIVE SUMMARY ............................................................................................. vi SECTION 1: INTRODUCTION TO BIOTRADE, -

Realizing the Potential of Synthetic Biology
Nature Reviews Molecular Cell Biology | AOP, published online 12 March 2014; doi:10.1038/nrm3767 PERSPECTIVES computer-aided-design (CAD), safety sys- VIEWPOINT tems, integrating models, genome editing and accelerated evolution. Synthetic biology Realizing the potential of synthetic is less like highly modular (or ‘switch-like’) electrical engineering and computer science biology and more like civil and mechanical engineer- ing in its use of optimization of modelling of whole system-level stresses and traffic flow. George M. Church, Michael B. Elowitz, Christina D. Smolke, Christopher A. Voigt and Ron Weiss Michael B. Elowitz. At the most general level, synthetic biology expands the subject Abstract | Synthetic biology, despite still being in its infancy, is increasingly matter of biology from the (already enor- providing valuable information for applications in the clinic, the biotechnology mous) space of existing species and cellular industry and in basic molecular research. Both its unique potential and the systems that have evolved to the even larger challenges it presents have brought together the expertise of an eclectic group of space of non-natural, but feasible, species scientists, from cell biologists to engineers. In this Viewpoint article, five experts and systems. Although we started with circuits to carry out the simplest kinds of discuss their views on the future of synthetic biology, on its main achievements in dynamic behaviours, synthetic approaches basic and applied science, and on the bioethical issues that are associated with the can be applied broadly to all types of bio design of new biological systems. logical functions from metabolism to multi cellular development. Synthetic biology allows us to figure out what types of genetic An increasing number of publications For instance, insight gained from systems circuit designs are capable of implement- and institutions are dedicated to biology investigations of natural processes ing different cellular behaviours, and what synthetic biology. -

2018 Semiconductor Synthetic Biology Roadmap
2018 Semiconductor Synthetic Biology Roadmap Editor’s Note Victor Zhirnov Chief Scientist at Semiconductor I am delighted to introduce the 1st Edition of the SemiSynBio Roadmap, a collective Research Corporation and Editor of work by many dedicated contributors from industry, academia and government. the 1st Edition of the Semiconductor It can be argued that innovation explosions often occur at the intersection of Roadmap. scientific disciplines, and Semiconductor Synthetic Biology or SemiSynBio is an excellent example of this. The objective of this Roadmap is to serve as a vehicle to realize the transformative potential of the new technology emerging at the interface between semiconductors and synthetic biology. The SemiSynBio Roadmap is intended to catalyze both interest in and rapid technological advances that provide new capabilities that benefit humankind. Victor Zhirnov is Chief Scientist at the Semiconductor Research Corporation. His research interests include nanoelectronics devices and systems, properties of materials at the nanoscale, bio-inspired electronic systems etc. He has authored and co-authored over 100 technical papers and contributions to books. Victor Zhirnov served as the Chair of the Emerging Research Device (ERD) Working Group for the International Technology Roadmap for Semiconductors (ITRS). Victor Zhirnov also holds adjunct faculty position at North Carolina State University and has served as an advisor to a number of government, industrial, and academic institutions. Victor Zhirnov received the M.S. in applied physics from the Ural Polytechnic Institute, Ekaterinburg, Russia, and the Ph.D. in solid state electronics and microelectronics from the Institute of Physics and Technology, Moscow, in 1989 and 1992, respectively. From 1992 to 1998 he was a senior scientist at the Institute of Crystallography of Russian Academy of Science in Moscow. -
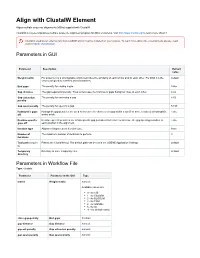
Align with Clustalw Element
Align with ClustalW Element Aligns multiple sequence alignments (MSAs) supplied with ClustalW. ClustalW is a general purpose multiple sequence alignment program for DNA or proteins. Visit http://www.clustal.org/ to learn more about it. Clustal is used as an external tool from UGENE and it must be installed on your system. To learn more about the external tools, please, read main UGENE User Manual. Parameters in GUI Parameter Description Default value Weight matrix For proteins it is a scoring table which describes the similarity of each amino acid to each other. For DNA it is the default scores assigned to matches and mismatches. End gaps The penalty for closing a gap. False Gap distance The gap separation penalty. Tries to decrease the chances of gaps being too close to each other. 4.42 Gap extension The penalty for extending a gap. 8.52 penalty Gap open penalty The penalty for opening a gap. 53.90 Hydrophilic gaps Hydrophilic gap penalties are used to increase the chances of a gap within a run (5 or more residues) of hydrophilic False off amino acids. Residue-specific Residue-specific penalties are amino specific gap penalties that reduce or increase the gap opening penalties at False gaps off each position in the alignment. Iteration type Alignment improvement iteration type. None Number of The maximum number of iterations to perform. 3 iterations Tool path (require Path to the ClustalW tool. The default path can be set in the UGENE Application Settings. default d) Temporary Directory to store temporary files. default directory Parameters -

Genocad Introduction
GenoCAD Introduction Computer-Assisted Design Software for Synthetic Biology Materials prepared by: Mary E. Mangan PhD www.openhelix.com/genocad Version 1 GenoCAD Introduction Agenda Introduction to GenoCAD genocad.org Introduction and Credits Register and Log In Step 1: Parts Parts Grammar Import the Training Set Step 2: Design Step 3: Simulate Summary Exercises GenoCAD: http://www.genocad.org/ Computer-assisted design software for synthetic biology Visit GenoCAD.org 1 Support for GenoCAD Process Flow click peccoud.org nsf.gov Parts and grammars, public or custom collections Peccoud team Design constructs NSF support Simulate processes Conceptual Framework for GenoCAD Data Model, Concepts Promoters Coding Seqs Terminators peccoud.vbi.vt.edu/publications 2007: 10.1093/bioinformatics/btm446 2009: 10.1371/journal.pcbi.1000529 Promoter A Coding Seq A Promoter B Coding Seq B Your constructs Parts form the foundation, stored in project libraries Aspects of DNA function explained with language metaphors: transcription, translation, code Grammar rules specify the way the parts work in series GenoCAD lets you develop language for programming cells Parts + Grammars give you synthetic construct designs 2 Further Reading Getting Assistance doi: 10.1093/bioinformatics/btm446 doi: 10.1371/journal.pcbi.1000529 doi: 10.1093/nar/gkp361 doi: 10.1016/j.tibtech.2011.09.001 support Help: context-sensitive help from ? Button Publications: theoretical, software, ongoing development Support link for asking questions or offering feedback -

Computational Tools and Algorithms for Designing Customized
View metadata, citation and similar papers at core.ac.uk brought to you by CORE REVIEW ARTICLE published: 06provided October by 2014 Frontiers - Publisher Connector BIOENGINEERING AND BIOTECHNOLOGY doi: 10.3389/fbioe.2014.00041 Computational tools and algorithms for designing customized synthetic genes Nathan Gould 1, Oliver Hendy 2 and Dimitris Papamichail 1* 1 Department of Computer Science, The College of New Jersey, Ewing, NJ, USA 2 Department of Biology, The College of New Jersey, Ewing, NJ, USA Edited by: Advances in DNA synthesis have enabled the construction of artificial genes, gene cir- Ilias Tagkopoulos, University of cuits, and genomes of bacterial scale. Freedom in de novo design of synthetic constructs California, Davis, USA provides significant power in studying the impact of mutations in sequence features, and Reviewed by: verifying hypotheses on the functional information that is encoded in nucleic and amino Yinjie Tang, Washington University, USA acids. To aid this goal, a large number of software tools of variable sophistication have M. Kalim Akhtar, University College been implemented, enabling the design of synthetic genes for sequence optimization London, UK based on rationally defined properties. The first generation of tools dealt predominantly *Correspondence: with singular objectives such as codon usage optimization and unique restriction site incor- Dimitris Papamichail, Department of Computer Science, The College of poration. Recent years have seen the emergence of sequence design tools that aim to New Jersey, 2000 Pennington Road, evolve sequences toward combinations of objectives.The design of optimal protein-coding Ewing, NJ 08628, USA sequences adhering to multiple objectives is computationally hard, and most tools rely on e-mail: [email protected] heuristics to sample the vast sequence design space. -

Bioinformatics in Metabolic Engineering
International Journal of Research Studies in Biosciences (IJRSB) Volume 7, Issue 11, 2019, PP 10-13 ISSN No. (Online) 2349-0365 DOI: http://dx.doi.org/10.20431/2349-0365.0711002 www.arcjournals.org Bioinformatics in Metabolic Engineering Nida Tabassum Khan*, Namra Jameel, Muhammad Jibran Khan Department of Biotechnology, Faculty of Life Sciences and Informatics, Balochistan University of Information Technology Engineering and Management Sciences, (BUITEMS), Quetta, Pakistan *Corresponding Author: Nida Tabassum Khan, Department of Biotechnology, Faculty of Life Sciences and Informatics, Balochistan University of Information Technology Engineering and Management Sciences, (BUITEMS), Quetta, Pakistan. Abstract: The emergence of metabolic engineering is strengthened by the tools of bioinformatics that not only supports experimental techniques employed for the manipulation of biological pathways but also provides an insilco platform for their study. There are numerous bioinformatics tools and software such as KEGG, MetaCyc,PH USER, Cytoscape etc that could be utilized by researchers in effectively transforming cellular pathways. Keywords: Systems biology ontology; Metabolic pathways; Gene network; Network visualization 1. INTRODUCTION Metabolic engineering integrates efficient exploration of metabolic and cellular pathways with molecular systems to enhance biological properties by formulating and applying genetic alterations [1].Decades ago, computational tools were used in metabolic engineering to design single enzymes by using analog computers, and digital computers were used to design the dynamics of metabolic pathways [2]. In recent years, bioinformatics tools are used to search huge metabolic databases leading to better understanding, managing, analyzing and visualizing of metabolic pathways [3, 4]. 2. APPLICATIONS OF BIOINFORMATICS TOOLS IN METABOLIC ENGINEERING There are several applications of bioinformatics tools in metabolic engineering which are as follows: 2.1. -

Real Time Collaboration on Multiple Sequence Alignments Jeffrey Yunes Submitted in Partial Satisfaction of the Require
ShareMSA: real time collaboration on multiple sequence alignments Jeffrey Yunes Submitted in partial satisfaction of the requirements for the degree of m in the GRADUATE DIVISION UNIVERSITY OF CALIFORNIA, SAN FRANCISCO ii Acknowledgements I am very thankful for the guidance and support of my thesis advisor and chair, Patricia C. Babbitt. I thank my thesis committee members Hao Li and Ian Holmes. I am grateful to Elaine Meng for feedback on user experience and Eric Pettersen for help with Chimera plugins. This work was funded by NIH GM60595 to Patricia C. Babbitt. iii Table of Contents Chapter1.................................................. 1 Abstract................................................. 1 Introduction............................................... 1 Features................................................. 1 ExamplesofUse ............................................ 2 Conclusion ............................................... 4 References . 5 iv List of Figures Figure 1 Screenshot from the mobile version of the site . 3 v Abstract A Multiple Sequence Alignment (MSA) is a widely used data structure in bioinformatics for applications ranging from evolutionary analyses to protein engineering. ShareMSA is a website that allows users to visualize, share, and collaborate on MSAs. ShareMSA makes MSAs more accessible by allowing collaborators one-click access to the MSA and annotations without the need to install software. In addition, when those with requisite permissions change the annotations or legend, all viewers of the MSA see the changes in real time. A Chimera plugin is available via the ShareMSA website, allowing Chimera users to publish MSAs from within this sequence-structure visualization software. Availability and Implementation ShareMSA is freely available at https://www.sharemsa.com, and does not require registration. The front end is implemented in HTML and JavaScript and runs on all modern browsers that support WebSockets.