Phylum Spirochaetes) Agustín Estrada-Peña1* and Alejandro Cabezas-Cruz2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Are You Suprised ?
B DAMB 721 Microbiology Final Exam B 100 points December 11, 2006 Your name (Print Clearly): _____________________________________________ I. Matching: The questions below consist of headings followed by a list of phrases. For each phrase select the heading that best describes that phrase. The headings may be used once, more than once or not at all. Mark the answer in Part 2 of your answer sheet. 1. capsid 7. CD4 2. Chlamydia pneumoniae 8. Enterococcus faecalis 3. oncogenic 9. hyaluronidase 4. pyruvate 10. interferon 5. Koplik’s spot 11. hydrophilic viruses 6. congenital Treponema pallidum 12. Streptococcus pyogenes 1. “spreading factor” produced by members of the staphylococci, streptococci and clostridia 2. viral protein coat 3. central intermediate in bacterial fermentation 4. persistant endodontic infections 5. a cause of atypical pneumonia 6. nonspecific defense against viral infection 7. has a rudimentary life cycle 8. HIV receptor 9. Hutchinson’s Triad 10. measles 11. resistant to disinfection 12. β-hemolytic, bacitracin sensitive, cause of suppurative pharyngitis 2 Matching (Continued): The questions below consist of diseases followed by a list of etiologic agents. Match each disease with the etiologic agent. Continue using Part 2 of your answer sheet. 1. dysentery 6. Legionnaire’s 2. botulism 7. gas gangrene 3. cholera 8. tuberculosis 4. diphtheria 9. necrotizing fascitis 5. enteric fever 10. pneumoniae/meningitis 13. Clostridium botulinum 14. Vibrio cholera 15. Mycobacterium bovis 16. Shigella species 17. Streptococcus pneumoniae 18. Clostridium perfringens 19. Salmonella typhi 20. Streptococcus pyogenes 3 II. Multiple Choice: Choose the ONE BEST answer. Mark the correct answer on Part 1 of the answer sheet. -

Differences in Genotype, Clinical Features, and Inflammatory
RESEARCH Differences in Genotype, Clinical Features, and Inflammatory Potential of Borrelia burgdorferi sensu stricto Strains from Europe and the United States Tjasa Cerar,1 Franc Strle,1 Dasa Stupica, Eva Ruzic-Sabljic, Gail McHugh, Allen C. Steere, Klemen Strle Borrelia burgdorferi sensu stricto isolates from patients with B. afzelii, which usually remains localized to the skin, with erythema migrans in Europe and the United States and B. garinii, which is usually associated with nervous were compared by genotype, clinical features of infection, system involvement (1). B. burgdorferi infection is rare in and inflammatory potential. Analysis of outer surface pro- Europe; little is known about its clinical course there. In tein C and multilocus sequence typing showed that strains the United States, B. burgdorferi is the sole agent of Lyme from these 2 regions represent distinct genotypes. Clinical borreliosis; in the northeastern United States, it is particu- features of infection with B. burgdorferi in Slovenia were similar to infection with B. afzelii or B. garinii, the other 2 larly arthritogenic (1,2). For all 3 species, the first sign of Borrelia spp. that cause disease in Europe, whereas B. infection is often an erythema migrans lesion. However, B. burgdorferi strains from the United States were associ- burgdorferi infection in the United States is associated with ated with more severe disease. Moreover, B. burgdorferi a greater number of disease-associated symptoms and more strains from the United States induced peripheral blood frequent hematogenous dissemination than B. afzelii or B. mononuclear cells to secrete higher levels of cytokines garinii infection in Europe (4–7). -

Association of Borrelia Garinii and B. Valaisiana with Songbirds in Slovakia
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Public Health Resources Public Health Resources 5-2003 Association of Borrelia garinii and B. valaisiana with Songbirds in Slovakia Klara Hanincova Department of Infectious Disease Epidemiology, Imperial College of Science, Technology and Medicine, London W2 1PG Veronika Taragelova Institute of Zoology, Slovak Academy of Science, 81364 Bratislava Juraj Koci Department of Biology, Microbiology and Immunology, University of Trnava, 918 43 Trnava, Slovakia Stefanie M. Schafer Department of Infectious Disease Epidemiology, Imperial College of Science, Technology and Medicine, London W2 1PG Rosie Hails NERC Centre of Ecology and Hydrology, Oxford OX 1 3SR See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/publichealthresources Part of the Public Health Commons Hanincova, Klara; Taragelova, Veronika; Koci, Juraj; Schafer, Stefanie M.; Hails, Rosie; Ullmann, Amy J.; Piesman, Joseph; Labuda, Milan; and Kurtenbach, Klaus, "Association of Borrelia garinii and B. valaisiana with Songbirds in Slovakia" (2003). Public Health Resources. 115. https://digitalcommons.unl.edu/publichealthresources/115 This Article is brought to you for free and open access by the Public Health Resources at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Public Health Resources by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Klara Hanincova, Veronika Taragelova, Juraj Koci, Stefanie M. Schafer, Rosie Hails, Amy J. Ullmann, Joseph Piesman, Milan Labuda, and Klaus Kurtenbach This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ publichealthresources/115 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, May 2003, p. 2825–2830 Vol. 69, No. 5 0099-2240/03/$08.00ϩ0 DOI: 10.1128/AEM.69.5.2825–2830.2003 Copyright © 2003, American Society for Microbiology. -

Pronunciation Guide to Microorganisms
Pronunciation Guide to Microorganisms This pronunciation guide is provided to aid each student in acquiring a greater ease in discussing, describing, and using specific microorganisms. Please note that genus and species names are italicized. If they cannot be italicized, then they should be underlined (example: a lab notebook). Prokaryotic Species Correct Pronunciation Acetobacter aceti a-se-toh-BAK-ter a-SET-i Acetobacter pasteurianus a-se-toh-BAK-ter PAS-ter-iann-us Acintobacter calcoacetius a-sin-ee-toe-BAK-ter kal-koh-a-SEE-tee-kus Aerococcus viridans (air-o)-KOK-kus vi-ree-DANS Agrobacterium tumefaciens ag-roh-bak-TEAR-ium too-me-FAY-she-ens Alcaligenes denitrificans al-KAHL-li-jen-eez dee-ni-TREE-fee-cans Alcaligenes faecalis al-KAHL-li-jen-eez fee-KAL-is Anabaena an-na-BEE-na Azotobacter vinelandii a-zoe-toe-BAK-ter vin-lan-DEE-i Bacillus anthracis bah-SIL-lus AN-thray-sis Bacillus lactosporus bah-SIL-lus LAK-toe-spore-us Bacillus megaterium bah-SIL-lus Meg-a-TEER-ee-um Bacillus subtilis bah-SIL-lus SA-til-us Borrelia recurrentis bore-RELL-ee-a re-kur-EN-tis Branhamella catarrhalis bran-hem-EL-ah cat-arr-RAH-lis Citrobacter freundii sit-roe-BACK-ter FROND-ee-i Clostridium perfringens klos-TREH-dee-um per-FRINGE-enz Clostridium sporogenes klos-TREH-dee-um spore-AH-gen-ease Clostridium tetani klos-TREH-dee-um TET-ann-ee Corynebacterium diphtheriae koh-RYNE-nee-back-teer-ee-um dif-THEE-ry-ee Corynebacterium hofmanni koh-RYNE-nee-back-teer-ee-um hoff-MAN-eye Corynebacterium xerosis koh-RYNE-nee-back-teer-ee-um zer-OH-sis Enterobacter -

View Tickborne Diseases Sample Report
1360 Bayport Ave, Ste B. San Carlos, CA 94070 1(866) 364-0963 | [email protected] | www. vibrant-wellness.com PATIENT PROVIDER NAME: DEMO REPORT GENDER: Male PRACTICE NAME: Vibrant IT4 Practice DATE OF BIRTH: 04/14/1998 AGE: 22 PROVIDER NAME: Demo Client, DDD (999994) ADDRESS: TEST STREET, TEST CITY, KY- 42437. ACCESSION ID: 2009220006 PHLEBOTOMIST: 607 SPECIMEN COLLECTION TIME: 09-21-2020 11:14 SPECIMEN RECEIVED TIME: 09-22-2020 05:14 FINAL REPORT TIME: 09-25-2020 15:56 FASTING: FASTING Your Vibrant Wellness TickBorne 2.0 panel results are enclosed. These results are intended to aid in the diagnosis of tickborne diseases by your healthcare provider. The Vibrant Tickborne Diseases panel tests for IgG and IgM antibodies for Borreliosis/Lyme disease as well as co-infection(s) and opportunistic infections with other tick-borne illnesses along with detection of DNA of the species causing these infections. The Vibrant Immunochip test is a semiquantitative assay that detects IgG and IgM antibodies in human serum. The PCR Test is a real-time PCR Assay designed for qualitative detection of infectious group- specific DNA in clinical samples. Interpretation of Report: The test results of antibody levels to the individual antigens are calculated by comparing the average intensity of the individual antibody to that of a reference population and cut-off chosen for each protein. Reference ranges have been established using a well characterized set of more than 300 serum samples and antibodies to specific bacteria tested. The results are displayed as In Control, Moderate, or High Risk.for each antigen tested. -
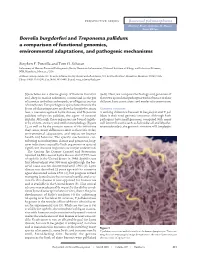
Borrelia Burgdorferi and Treponema Pallidum: a Comparison of Functional Genomics, Environmental Adaptations, and Pathogenic Mechanisms
PERSPECTIVE SERIES Bacterial polymorphisms Martin J. Blaser and James M. Musser, Series Editors Borrelia burgdorferi and Treponema pallidum: a comparison of functional genomics, environmental adaptations, and pathogenic mechanisms Stephen F. Porcella and Tom G. Schwan Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, NIH, Hamilton, Montana, USA Address correspondence to: Tom G. Schwan, Rocky Mountain Laboratories, 903 South 4th Street, Hamilton, Montana 59840, USA. Phone: (406) 363-9250; Fax: (406) 363-9445; E-mail: [email protected]. Spirochetes are a diverse group of bacteria found in (6–8). Here, we compare the biology and genomes of soil, deep in marine sediments, commensal in the gut these two spirochetal pathogens with reference to their of termites and other arthropods, or obligate parasites different host associations and modes of transmission. of vertebrates. Two pathogenic spirochetes that are the focus of this perspective are Borrelia burgdorferi sensu Genomic structure lato, a causative agent of Lyme disease, and Treponema A striking difference between B. burgdorferi and T. pal- pallidum subspecies pallidum, the agent of venereal lidum is their total genomic structure. Although both syphilis. Although these organisms are bound togeth- pathogens have small genomes, compared with many er by ancient ancestry and similar morphology (Figure well known bacteria such as Escherichia coli and Mycobac- 1), as well as by the protean nature of the infections terium tuberculosis, the genomic structure of B. burgdorferi they cause, many differences exist in their life cycles, environmental adaptations, and impact on human health and behavior. The specific mechanisms con- tributing to multisystem disease and persistent, long- term infections caused by both organisms in spite of significant immune responses are not yet understood. -

Transhemispheric Exchange of Lyme Disease Spirochetes by Seabirds BJO¨ RN OLSEN,1,2 DAVID C
JOURNAL OF CLINICAL MICROBIOLOGY, Dec. 1995, p. 3270–3274 Vol. 33, No. 12 0095-1137/95/$04.0010 Copyright q 1995, American Society for Microbiology Transhemispheric Exchange of Lyme Disease Spirochetes by Seabirds BJO¨ RN OLSEN,1,2 DAVID C. DUFFY,3 THOMAS G. T. JAENSON,4 ÅSA GYLFE,1 1 1 JONAS BONNEDAHL, AND SVEN BERGSTRO¨ M * Department of Microbiology1 and Department of Infectious Diseases,2 Umeå University, S-901 87 Umeå, and Department of Zoology, Section of Entomology, and Zoological Museum, University of Uppsala, S-752 36 Uppsala,4 Sweden, and Alaska Natural Heritage Program, Environment and Natural Resources Institute, University of Alaska, Anchorage, Anchorage, Alaska 995013 Received 19 June 1995/Returned for modification 17 August 1995/Accepted 18 September 1995 Lyme disease is a zoonosis transmitted by ticks and caused by the spirochete Borrelia burgdorferi sensu lato. Epidemiological and ecological investigations to date have focused on the terrestrial forms of Lyme disease. Here we show a significant role for seabirds in a global transmission cycle by demonstrating the presence of Lyme disease Borrelia spirochetes in Ixodes uriae ticks from several seabird colonies in both the Southern and Northern Hemispheres. Borrelia DNA was isolated from I. uriae ticks and from cultured spirochetes. Sequence analysis of a conserved region of the flagellin (fla) gene revealed that the DNA obtained was from B. garinii regardless of the geographical origin of the sample. Identical fla gene fragments in ticks obtained from different hemispheres indicate a transhemispheric exchange of Lyme disease spirochetes. A marine ecological niche and a marine epidemiological route for Lyme disease borreliae are proposed. -

Information to Users
INFORMATION TO USERS This manuscript bas been reproJuced from the microfilm master. UMI films the text directly ftom the original or copy submitted. Thus, sorne thesis and dissertation copies are in typewriter face, while others may be itom any type ofcomputer printer. The quality oftbis reproduction is depeDdeDt apoD the quality of the copy sablDitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthlough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will he noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand corner and continuing trom left to right in equal sections with sma1l overlaps. Each original is a1so photographed in one exposure and is included in reduced fonn at the back orthe book. Photographs ineluded in the original manuscript have been reproduced xerographically in this copy. Higher quality 6" x 9" black and white photographie prints are available for any photographs or illustrations appearing in this copy for an additional charge. Contact UMI directly to order. UMI A Bell & Howell Information Company 300 North Zeeb Raad, ADn AJbor MI 48106-1346 USA 313n61-4700 8OO1S21~ NOTE TO USERS The original manuscript received by UMI contains pages with slanted print. Pages were microfilmed as received. This reproduction is the best copy available UMI Oral spirochetes: contribution to oral malodor and formation ofspherical bodies by Angela De Ciccio A thesis submitted to the Faculty ofGraduate Studies and Research, McGill University, in partial fulfillment ofthe requirements for the degree ofMaster ofScience. -

First Cases of Natural Infections with Borrelia Hispanica in Two Dogs and a Cat from Europe
microorganisms Case Report First Cases of Natural Infections with Borrelia hispanica in Two Dogs and a Cat from Europe 1, , 2, 2 3 3 Gabriele Margos * y, Nikola Pantchev y, Majda Globokar , Javier Lopez , Jaume Rodon , Leticia Hernandez 3, Heike Herold 4 , Noelia Salas 3, Anna Civit 3 and Volker Fingerle 1 1 German National Reference Centre for Borrelia, Bavarian Health and Food Safety Authority, 85764 Oberschleißheim, Germany; volker.fi[email protected] 2 IDEXX Laboratories, 70806 Kornwestheim, Germany; [email protected] (N.P.); [email protected] (M.G.) 3 IDEXX Laboratories, 08038 Barcelona, Spain; [email protected] (J.L.); [email protected] (J.R.); [email protected] (L.H.); [email protected] (N.S.); [email protected] (A.C.) 4 Bavarian Health and Food Safety Authority, 85764 Oberschleißheim, Germany; [email protected] * Correspondence: [email protected] These authors contributed equally to this work. y Received: 21 July 2020; Accepted: 14 August 2020; Published: 18 August 2020 Abstract: Canine cases of relapsing fever (RF) borreliosis have been described in Israel and the USA, where two RF species, Borrelia turicatae and Borrelia hermsii, can cause similar clinical signs to the Borrelia persica in dogs and cats reported from Israel, including fever, lethargy, anorexia, thrombocytopenia, and spirochetemia. In this report, we describe the first clinical cases of two dogs and a cat from Spain (Cordoba, Valencia, and Seville) caused by the RF species Borrelia hispanica. Spirochetes were present in the blood smears of all three animals, and clinical signs included lethargy, pale mucosa, anorexia, cachexia, or mild abdominal respiration. -

The Relapsing Fever Agent Borrelia Hermsii Has Multiple
Copyright Q 1992 by the Genetics Societyof America The Relapsing Fever AgentBorrelia hermsii Has Multiple Copiesof Its Chromosome and Linear Plasmids Todd Kitten and Alan G. Barbour Departments of Microbiology and Medicine, The Universityof Texas Health Science Center, Sun Antonio, Texas 78284 Manuscript received March 17, 1992 Accepted for publicationJune 25, 1992 ABSTRACT Borrelia hermsii, a spirochete which causes relapsing fever in humans and other mammals, eludes the immune responseby antigenic variationof the “Vmp” proteins.This occurs by replacement of an expressed vrnp gene with a copy of a silent vrnp gene. Silent and expressedvrnp genes are located on separate linearplasmids. To further characterize vrnp recombination, copy numbers were determined for two linear plasmids and for the l-megabase chromosome by comparing hybridization of probes to native DNA with hybridization to recombinant plasmids containing borrelial DNA. Plasmid copy numbers were also estimated by ethidium bromide fluorescence. Total cellular DNA content was determined by spectrophotometry. For borreliasgrown in mice, copy numbers and 95% confidence intervals were 14 (12-17)for an expression plasmid,8 (7-9) for a silent plasmid, and 16 (13-18) for the chromosome. Borrelias grown in broth mediumhad one-fourth to one-half this number of plasmids and chromosomes. Staining of cells with4’,6-diamidino-2-phenylindole revealed DNA to be distributed throughout most of the spirochete’s length. These findings indicate that borrelias organize their total cellular DNA into several complete genomes and thatcells undergoing serotype switches do one or more of the following: (1) coexpress Vmps from switched and unswitched expression plasmids for at least three to five generations, (2)suppress transcription from some expressionplasmid copies, or (3) partition expression plasmids nonrandomly.The lower copy number ofthe silentplasmid indicates that nonreciprocal Vmp gene recombination may result from loss of recombinant silent ” plasmids by segregation. -

Taxonomy JN869023
Species that differentiate periods of high vs. low species richness in unattached communities Species Taxonomy JN869023 Bacteria; Actinobacteria; Actinobacteria; Actinomycetales; ACK-M1 JN674641 Bacteria; Bacteroidetes; [Saprospirae]; [Saprospirales]; Chitinophagaceae; Sediminibacterium JN869030 Bacteria; Actinobacteria; Actinobacteria; Actinomycetales; ACK-M1 U51104 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Limnohabitans JN868812 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae JN391888 Bacteria; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Planctomyces HM856408 Bacteria; Planctomycetes; Phycisphaerae; Phycisphaerales GQ347385 Bacteria; Verrucomicrobia; [Methylacidiphilae]; Methylacidiphilales; LD19 GU305856 Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; Pelagibacteraceae GQ340302 Bacteria; Actinobacteria; Actinobacteria; Actinomycetales JN869125 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae New.ReferenceOTU470 Bacteria; Cyanobacteria; ML635J-21 JN679119 Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae HM141858 Bacteria; Acidobacteria; Holophagae; Holophagales; Holophagaceae; Geothrix FQ659340 Bacteria; Verrucomicrobia; [Pedosphaerae]; [Pedosphaerales]; auto67_4W AY133074 Bacteria; Elusimicrobia; Elusimicrobia; Elusimicrobiales FJ800541 Bacteria; Verrucomicrobia; [Pedosphaerae]; [Pedosphaerales]; R4-41B JQ346769 Bacteria; Acidobacteria; [Chloracidobacteria]; RB41; Ellin6075 -

Proteome Characterization of Brachyspira Strains
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Department of Cellular Biology, Physiology and Immunology Doctoral Program in Immunology Proteome characterization of Brachyspira strains. Identification of bacterial antigens. Doctoral Thesis Mª Vanessa Casas López Bellaterra, July 2017 Department of Cellular Biology, Physiology and Immunology Doctoral Program in Immunology Proteome characterization of Brachyspira strains. Identification of bacterial antigens. Doctoral thesis presented by Mª Vanessa Casas López To obtain the Ph.D. in Immunology This work has been carried out in the Proteomics Laboratory CSIC/UAB under the supervision of Dr. Joaquin Abián and Dra. Montserrat Carrascal. Ph.D. Candidate Ph.D. Supervisor Mª Vanessa Casas López Dr. Joaquin Abián Moñux CSIC Research Scientist Department Tutor Ph.D. Supervisor Dra. Dolores Jaraquemada Pérez de Dra. Montserrat Carrascal Pérez Guzmán CSIC Tenured Scientist UAB Immunology Professor Bellaterra, July 2017 “At My Most Beautiful” R.E.M. from the album “Up” (1998) “And after all, you’re my wonderwall” Oasis from the album “(What´s the Story?) Morning Glory” (1995) Agradecimientos A mis directores de tesis, por su tiempo, sus ideas y consejos.