Package 'Ade4'
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Multivariate Statistical Analysis of Decathlon Performance Results in Olympic Athletes (1988-2008)
World Academy of Science, Engineering and Technology International Journal of Sport and Health Sciences Vol:5, No:5, 2011 Multivariate Statistical Analysis of Decathlon Performance Results in Olympic Athletes (1988-2008) Jaebum Park, and Vladimir M. Zatsiorsky interdependency between the decathlon events from a more Abstract—The performance results of the athletes competed in general, biological, perspective. Their interest was in testing the 1988-2008 Olympic Games were analyzed (n = 166). The data the so-called ‘principle of allocation’ which states that the were obtained from the IAAF official protocols. In the principal physical performance by vertebrates is strongly influenced by component analysis, the first three principal components explained tradeoffs between the pairs of ecologically important features 70% of the total variance. In the 1st principal component (with 43.1% of total variance explained) the largest factor loadings were and between conflicting specialist and generalist phenotypes for 100m (0.89), 400m (0.81), 110m hurdle run (0.76), and long jump [4]. The authors constructed a data set of the performance of (–0.72). This factor can be interpreted as the ‘sprinting performance’. 600 world-class decathletes. Similar to the previous findings The loadings on the 2nd factor (15.3% of the total variance) [6] individual performance for any pairs of disciplines was presented a counter-intuitive throwing-jumping combination: the positively correlated. The authors considered this fact highest loadings were for throwing events (javelin throwing 0.76; ‘unexpected’. According to their opinion, the above shot put 0.74; and discus throwing 0.73) and also for jumping events (high jump 0.62; pole vaulting 0.58). -

Preserving Fabled Amateurism: the Benefits of the NCAA's Adoption of the Olympic Amateurism Model
Journal of Law and Policy Volume 29 Issue 1 Article 8 12-1-2020 Preserving Fabled Amateurism: The Benefits of the NCAA’s Adoption of the Olympic Amateurism Model John Kealey Follow this and additional works at: https://brooklynworks.brooklaw.edu/jlp Part of the Antitrust and Trade Regulation Commons, Contracts Commons, Education Law Commons, Entertainment, Arts, and Sports Law Commons, First Amendment Commons, Intellectual Property Law Commons, and the Labor and Employment Law Commons Recommended Citation John Kealey, Preserving Fabled Amateurism: The Benefits of the NCAA’s Adoption of the Olympic Amateurism Model, 29 J. L. & Pol'y 325 (2020). Available at: https://brooklynworks.brooklaw.edu/jlp/vol29/iss1/8 This Note is brought to you for free and open access by the Law Journals at BrooklynWorks. It has been accepted for inclusion in Journal of Law and Policy by an authorized editor of BrooklynWorks. PRESERVING FABLED AMATEURISM: THE BENEFITS OF THE NCAA’S ADOPTION OF THE OLYMPIC AMATEURISM MODEL John Kealey* I’m not only saying that it is a right for a [collegiate athlete] to play summer ball for money . but I’m going further than that. [They are] failing in [their] duty to [them]self and to the world if [they do] not take advantage of it and use it to the best of [their] ability. – G. Stanley Hall, President, Clark University1 After a century of denying student-athletes from receiving compensation outside the cost of attendance for their athletic contributions to their respective universities, the NCAA finally announced it would change its amateurism rule. The change came in response to multiple class action lawsuits and, more recently, legislation from many states, namely California and New York, which would have mandated that universities do not interfere with student-athletes desire to commercially exploit their own names, image, and likenesses. -

Bob Mathias: Olympian and Football Player
VOL. XX, NO. III MAY 2007 BOB MATHIAS OLYMPIAN AND FOOTBALL PLAYER By Keith McClellan The recent death of Olympic decathlon champion Robert Bruce Mathias reminds many college football fans that Bob Mathias is the only man ever to participate in both the Rose Bowl and the Olympic Games in the same year (Herman Brix won a silver medal in the shotput at the 1928 Olympics but his Rose Bowl appearance as a left tackle for Washington came in the 1926 Rose Bowl game). Given the hyperbolae about his political success, back-to-back wins in the Olympic decathlon -- with a world record performance in 1952 - the Sullivan Award as the nation's outstanding amateur athlete, and a leading role in a Hollywood movie about himself, it is easy to forget that he was a great college football player. On November 17,1930, Bob became the second of four children born to Charles and Lillian Mathias, a family doctor and his wife, in Tulare, California, a small farming town of 12,000 people in the San Joaquin Valley. He entered Tulare Union High School as a freshman during World War II in the fall of 1944. Bob was a natural athlete, lifted two hundred pound bags of insecticide aboard crop dusting planes in the summer, and lettered all four years for the Tulare Redskins in basketball and track He was not allowed to play football as a high school sophomore because, according to the California Interscholastic Federation rules, he was too heavy to play in Class B football and too young to play in Class A. -

Developing a Formula for the Comparison of Athletics Performances Across Gender, Age and Event Boundaries Based on South African Standards
Vaal University of Technology Developing a formula for the comparison of athletics performances across gender, age and event boundaries based on South African standards SWJ Bekker (083 734-7079) 212118781 Dissertation Magister Technologiae: Information Technology In the FACULTY OF APPLIED AND COMPUTER SCIENCES Department of Information and Communication Technology At the Vaal University of Technology Supervisor: Dr J. F. Janse van Rensburg Declaration I, Sarel Wilhelmus Jacobus Bekker, ID number 4109185061087, hereby declare that the content of this dissertation is my own unaided work and that all quoted in this dissertation is properly acknowledged and referenced. Signed: ___________________ At Vanderbijlpark on 28/01/2018 i Acknowledgements Firstly and with humility, to GOD almighty for the ability and insight to conduct this work. To my departed wife who had to listen to my ideas and her support during the whole process of investigation, reading and writing. I wish to thank her mainly for being there for me as a soundboard when needed. To my daughters who are both involved in sport and have given me support and encouragement to continue. I want to thank Richard Stander (formerly from ASA and now with Boland Athletics) who has continually provided me with insight and information since 1985. I would also like to thank all the friends I have made through athletics, some only known by voice and a large number in person. Their continuous support (and criticism) has inspired me to continue with this work. My colleagues at Vaal University of Technology (especially Willem and Hannes) for their motivation to continue. Dr Pieter Conradie for giving initial direction to this work. -

Enter Issue 09 1984 Jul Aug.Pdf
, •••••••••••• p ••• Atari®presents the five greatest advances in the creative arts since someone put 72 crayons in one box. What would Cezanne say to an bois as well as words. So everyone basics you'll be ready to move up to electronic orange? Surely Van from preschoolers to grandparents ATARI Music Compose z4' and Gogh would go for some flowers can create without going near the create original compositions in four painted in phosphors (those glow keyboard. part harmony! ing things in your 1V screen). And All of these programs were de you bet Beethoven would be blown signed to get the best from your away by a computer synthesized ATARI Computer, including the symphony ATARI 800XL H ' or the less expensive Too bad. They were all born too ATAR! 600XU" Both machines give early But luckily you weren't. Be you unsurpassed Atari graphics cause Atari makes several home and four sound channels. And computer products to help you whether you're painting with light create all these things and more. or composing at the com- ~ First, there's ATARI Paint;" the puter keyboard, you can store program that turns the joystick you your creation on the ATARI E!P... already own into a computerized 101O T'I Program Recorder d paintbrush that helps you explore or the more sophisticated ~ H the fascinating world of computer 1050 ' Disk Drive. art. \ And if all that doesn't Get the muglc touch with Atart Light Pen lets you convince you that our new Atart Touch Tablet. write rtght on the screen. -
Jim Thorpe Chooses the Bright Path of Responsibility
Jim Thorpe Chooses the Bright Path of Responsibility Handout A: Narrative BACKGROUND The greatest achievement and most honored title in sports competition goes to the winner of the Olympic Decathlon. The Decathlon (a Greek word meaning “Ten Feats”) is the ultimate all-around test of athletic skill and discipline. It is a 10-event contest lasting two days that tests a wide range of athletic abilities. The first day consists of (in order): 100m race, Long Jump, Shot Put, High Jump, and 400m. The second day’s events are 110m Hurdles, Discus Throw, Pole Vault, Javelin Throw, and 1500m. The competitor with the most points in the ten events is the winner and is crowned with the title of “The World’s Greatest Athlete.” The legendary Jim Thorpe won a gold medal in the first modern Olympic Decathlon, which was introduced in the 1912 Olympics Games at Stockholm, Sweden. In those same Olympic Games, Thorpe also won a gold medal for the Pentathlon competition, which included five events comprised of the long jump, discus, javelin, sprint, and wrestling. The Pentathlon, which is no longer included in the Olympic Games, was the forerunner of the modern Decathlon and dates back to the ancient Olympic Games. When Thorpe won gold medals in both the Decathlon and Pentathlon, King Gustaf V of Sweden personally awarded the medals to Thorpe while grabbing his hand and proclaiming, “Sir, you are the greatest athlete in the world!” Throughout his life, Thorpe understood that he had a responsibility to use his abilities to help those around him. -
Liste Des Jeux - Version 128Go
Liste des Jeux - Version 128Go Amstrad CPC 2542 Apple II 838 Apple II GS 588 Arcade 4562 Atari 2600 2271 Atari 5200 101 Atari 7800 52 Channel F 34 Coleco Vision 151 Commodore 64 7294 Family Disk System 43 Game & Watch 58 Gameboy 621 Gameboy Advance 951 Gameboy Color 502 Game Gear 277 GX4000 25 Lynx 84 Master System 373 Megadrive 1030 MSX 1693 MSX 2 146 Neo-Geo Pocket 9 Neo-Geo Pocket Color 81 Neo-Geo 152 N64 78 NES 1822 Odyssey 2 125 Oric Atmos 859 PC-88 460 PC-Engine 291 PC-Engine CD 4 PC-Engine SuperGrafx 97 Pokemon Mini 25 Playstation 123 PSP 2 Sam Coupé 733 Satellaview 66 Sega 32X 30 Sega CD 47 Sega SG-1000 64 SNES 1461 Sufami Turbo 15 Thompson TO6 125 Thompson TO8 82 Vectrex 75 Virtual Boy 24 WonderSwan 102 WonderSwan Color 83 X1 614 X68000 546 Total 32431 Amstrad CPC 1 1942 Amstrad CPC 2 2088 Amstrad CPC 3 007 - Dangereusement Votre Amstrad CPC 4 007 - Vivre et laisser mourir Amstrad CPC 5 007 : Tuer n'est pas Jouer Amstrad CPC 6 1001 B.C. - A Mediterranean Odyssey Amstrad CPC 7 10th Frame Amstrad CPC 8 12 Jeux Exceptionnels Amstrad CPC 9 12 Lost Souls Amstrad CPC 10 1943: The Battle of Midway Amstrad CPC 11 1st Division Manager Amstrad CPC 12 2 Player Super League Amstrad CPC 13 20 000 avant J.C. Amstrad CPC 14 20 000 Lieues sous les Mers Amstrad CPC 15 2112 AD Amstrad CPC 16 3D Boxing Amstrad CPC 17 3D Fight Amstrad CPC 18 3D Grand Prix Amstrad CPC 19 3D Invaders Amstrad CPC 20 3D Monster Chase Amstrad CPC 21 3D Morpion Amstrad CPC 22 3D Pool Amstrad CPC 23 3D Quasars Amstrad CPC 24 3d Snooker Amstrad CPC 25 3D Starfighter Amstrad CPC 26 3D Starstrike Amstrad CPC 27 3D Stunt Rider Amstrad CPC 28 3D Time Trek Amstrad CPC 29 3D Voicechess Amstrad CPC 30 3DC Amstrad CPC 31 3D-Sub Amstrad CPC 32 4 Soccer Simulators Amstrad CPC 33 4x4 Off-Road Racing Amstrad CPC 34 5 Estrellas Amstrad CPC 35 500cc Grand Prix 2 Amstrad CPC 36 7 Card Stud Amstrad CPC 37 720° Amstrad CPC 38 750cc Grand Prix Amstrad CPC 39 A 320 Amstrad CPC 40 A Question of Sport Amstrad CPC 41 A.P.B. -
2008 US Olympic Trials Decathlon Handbook
MEDIA GUIDE / HANDBOOK US OLYMPIC TEAM DECATHLON TRIALS and 89th NATIONAL CHAMPIONSHIPS Hayward Field University of Oregon Eugene, Oregon June 29-30, 2008 Frank Zarnowski DECA, The Decathlon Association www.decathlonusa.typepad.com 0 Table of Contents Section One: Background Information page 2 Time Schedule 2 Qualifying Procedures 2 List of Qualifiers 3 Web sites which will post results 4 TV Coverage 4 Section Two: Record Section US Olympic Trials Winners, 1912-2004 5 Adenda 5 Number of Competitors/Finishers by Year 6 Individual Event Records 6 Recent Meet Results (1996, 2000, 2004) 9 Section Three: Athlete’s Bios Mustafa Abdur-Rahim page 15 Jangy Addy 16 Jake Arnold 17 Chris Boyles 18 Joe Cebulski 19 Raven Cepeda 20 Bryan Clay 22 Joe Detmer 23 Ashton Eaton 24 Trey Hardee 25 Ryan Harlan 26 Chris Helwick 27 Brandon Hoskins 29 Ricky Moody 30 Mike Morrison 30 Ryan Olkowski 32 Tom Pappas 33 Chris Randolph 35 Chris Richardson 36 Paul Terek 37 PR Page 39 1 SECTION ONE: Basic Info: a) Time Schedule b) Qualifying procedures c) List of Qualifiers d) Web sites which will provide results e) TV coverage a… Time Schedule Sunday, June 29, 2008 Monday, June 30, 2008 10:00 am 100 meters 11:30 am 110 m Hurdles 10:50 Long Jump 12;20 pm Discus Noon Shot Put 2:35 Pole Vault 1:15 pm High Jump 4:45 Javelin 4:00 400 meters 8:35 1500 meters b) …..Qualifying Procedure "A" and "B" Standards A standard is 7900, B standard is 7600. Field limit is 18. -
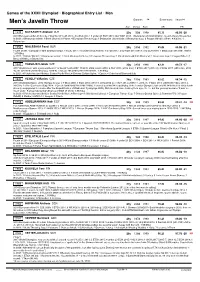
Men's Javelin Throw
Games of the XXXII Olympiad • Biographical Entry List • Men Men’s Javelin Throw Entrants: 34 Event starts: August 4 Age (Days) Born SB PB 1249 KATKAVETS Aliaksei BLR 23y 55d 1998 85.10 86.05 -20 2019 European under-23 bronze // dnq World Youth 2015; dnq EJC 2017; 3 under-23 ECH 2019; dnq WCH 2019. 1 Belarusian 2018/2020/2021. Coach-Valery Oksenchuk In 2021: 2 Belarusian winter; 1 Brest Shukevich Prizes; 4 European Throws Cup; 1 Belarusian Universiade; 2 Belarusian Cup; 2 Prague Odložil; 2 Brno; 4 Kladno; 1 Belarusian 1250 MIALESHKA Pavel BLR 28y 249d 1992 85.06 85.06 -21 Fourth at 2017 European Team Championships // 3 EJC 2011; 10 under-23 ECH 2013; 4 ETCh 2017; dnq WCH 2017/2019; dnq ECH 2018. 1 Belarusian 2013/2017/2019. 1.87/90kg In 2021: 1 Minsk ‘Winter’; 1 Belarusian winter; 2 Brest Shukevich Prizes; 3 European Throws Cup; 1 Minsk Romuald Klim Memorial; 1 Belarusian Cup; 1 Prague Odložil; 1 Brno; 5 Kladno; 2 Belarusian 1642 VADLEJCH Jakub CZE 30y 295d 1990 82.31 89.73 -17 2017 World silver, with a personal best // 12 World Youth 2007; 10 WJC 2008; 8 EJC 2009; 8 OLY 2016 (2012-dnq); 1 ETCh 2017 (2019-2); 2 WCH 2017 (2015-dnq, 2019 -5); 8 ECH 2018 (2010/2014-dnq, 2016-9). 1 Czech 2014/2015/2017/2019/2021. 1.91/93kg In 2021: 4 Potchefstroom Athletics Central North West; 4 Ostrava Golden Spike; 1 Czech; 4 Gateshead Diamond/July 1643 VESELÝ Vítězslav CZE 38y 155d 1983 82.63 88.34 -12 2013 World Champion, 2012 Olympic bronze // 9 WJC 2002; 1 ECH 2012 (2010-9, 2014/2016-2); 3 OLY 2012 (2008-11, 2016-7); 1 WCH 2013 (2009/2017-dnq, 2011-4, 2015-8); 2 IAAF Continental Cup 2014. -

Original Article Classification of Athletic Decathlon Using Methods of Hierarchical Analysis
Journal of Physical Education and Sport ® (JPES), Vol 20 (Supplement issue 6), Art 441 pp 3253 – 3259, 2020 online ISSN: 2247 - 806X; p-ISSN: 2247 – 8051; ISSN - L = 2247 - 8051 © JPES Original Article Classification of athletic decathlon using methods of hierarchical analysis JAROSLAV BROĎÁNI 1, NATÁLIA DVOŘÁČKOVÁ 2, MONIKA CZAKOVÁ 3 1,2,3, Department of Physical Education and Sport, Faculty of Education, Constantine the Philosopher University in Nitra, SLOVAKIA Published online: November 30, 2020 (Accepted for publication: November 22, 2020) DOI:10.7752/jpes.2020.s6441 Abstract Authors deals with the problematics of group classification of athletics disciplines, which influence the sports performance in the men's decathlon. For the group identification, the indicators of the best world's performance in decathlon above the 8000 points according to the data from IAAF (1986-2019) were used. From the classification methods of clustering the hierarchical models as the Average linkage (Between & Within-group), Single Linkage - Nearest neighbor, Complete Linkage - Farthest neighbor, Centroid linkage, Median clustering, and Ward´s method were used. The substructure of the clusters is differentiated according to the relations of used methods. All seven clustering methods agreed in four groups of disciplines in 4 cluster [100 meters, 400 meters, Long jump, 110 meters hurdles, Pole vault] [High jump] [1500 meters]. Stability test with a substructure of decathlon of 4th. cluster is identified in 85.71% cases. Hierarchical models allow identifying groups of athletics disciplines that influence the sports performance in men's decathlon. Understanding the structure of sports performance contributes to the streamlining of the training process and determining the typology of top-class decathletes. -

An Olympic Hurdle: Why Is the Decathlon Only for Men?
From Pentathlon to Decathlon, Pat Daniels-Winslow & Jordan Gray Pioneer Women’s Multi Events -- AT CSM – Over 6 Decades: This July 6, 2021, NEW YORK TIMES story features College of San Mateo’s 3-time Olympian (1960-64-68) Pat Daniels Winslow Connolly, who was national pentathlon champion (1961-67) and first to compete in the event in the Olympics (Tokyo 1964). The 1961 Capuchino High grad lives in Half Moon Bay. Also: Jordan Gray set the women’s decathlon American Record, winning the first national championship in 2019 -- held at CSM. Courtesy of the New York Times with lead-in by Fred Baer NEW YORK TIMES An Olympic Hurdle: Why Is the Decathlon Only for Men? Chasing opportunity and equality, women are campaigning for their own 10-event competition, rather than just the heptathlon, at the Paris Games in 2024. Competing as a heptathlete, Jordan Gray failed to qualify for the Tokyo Olympics at the U.S. track and field trials last month. Credit Alexandra Garcia/The New York Times By Shauna Farnell July 6, 2021Updated 9:16 a.m. ET Jordan Gray wants to bring a women’s decathlon to the Olympics. Even if it happens long after she is able to compete in the event. “The goal is 2024,” Gray, a 25-year-old Georgia native and the U.S. record-holder in the women’s decathlon, said recently by telephone. Her movement — Let Women Decathlon — is nearing 20,000 petition signatures in favor of adding the event to the Olympic Games in the name of gender equality in track and field, and it is gaining the support of Olympics icons who broke similar barriers decades ago. -

An Analysis of Elite Decathlon Performances
DOCUMENT MOVE ED 175 121 SP 014 469 _AUTHOR Freeman, Villiam H. TITLE An Analysis of Elite Decathlon Performances. POD DATE Mar 79 NOTE 13p.: Paper presented at Annual Meeting of the American Alliance for Health, Physical Education, Recreation and Dance (Mew Orleans, Louisiana, March 197r EDRS PRICE MF01/VC01 Plus Postage. DESCRIPTORS *Athletics: nomparative Analysis: Complexity Level: Correlation: Motor Development: *Performance: Physical Activities: *Scores: *Skill bevelopaent: Statistzcal Analysis IDENTIFIERS Decathlon ABSTRACT This analysis involved the ten events of the men's decathlon in all performances which scored 8,000 pointsor higher, based on the 1962 Scoring Tables of the International Amateur Athletic Federation. It attempted to (1) determine the interrelationships among the events and the finalscores,(2) look for areas of difference ccapared to sub-9,00C point performers, and (3) decide whether the performance levelscan be predicted on the basis of significantly fewer events. Coapared tc earlier perforsances, the results were sore balanced in relation to the scoring tables, and the correlationsamong and between the events and the final scores were generally lower thaa in previous studies. In addition, it yeses that elite-level athletes turn ina more balanced performance in their non-specialty events than do lesser coapetitors, and that in training the higher-level decathlete, early attention towards developdng all ten events equally towardsan optimal skill level should be encouraged. It also appears that theuse of predictive formulae are of little value when considering elite-level cospetition. (Author/LH) *********************************************************************** Reproductions supplied by EDRS are the best that can be made from the original document. *********************************************************************** Presented at Eesearch Section, American Alliance for Health, Physical Education and Recreation, New Orleans, LA, March 1979.