Matriarch: a Python Library for Materials Architecture † ‡ † ‡ § † ⊥ ‡ Tristan Giesa, , Ravi Jagadeesan, , , David I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department
Spoken Tutorial Project, IIT Bombay Brochure for Chemistry Department Name of FOSS Applications Employability GChemPaint GChemPaint is an editor for 2Dchem- GChemPaint is currently being developed ical structures with a multiple docu- as part of The Chemistry Development ment interface. Kit, and a Standard Widget Tool kit- based GChemPaint application is being developed, as part of Bioclipse. Jmol Jmol applet is used to explore the Jmol is a free, open source molecule viewer structure of molecules. Jmol applet is for students, educators, and researchers used to depict X-ray structures in chemistry and biochemistry. It is cross- platform, running on Windows, Mac OS X, and Linux/Unix systems. For PG Students LaTeX Document markup language and Value addition to academic Skills set. preparation system for Tex typesetting Essential for International paper presentation and scientific journals. For PG student for their project work Scilab Scientific Computation package for Value addition in technical problem numerical computations solving via use of computational methods for engineering problems, Applicable in Chemical, ECE, Electrical, Electronics, Civil, Mechanical, Mathematics etc. For PG student who are taking Physical Chemistry Avogadro Avogadro is a free and open source, Research and Development in Chemistry, advanced molecule editor and Pharmacist and University lecturers. visualizer designed for cross-platform use in computational chemistry, molecular modeling, material science, bioinformatics, etc. Spoken Tutorial Project, IIT Bombay Brochure for Commerce and Commerce IT Name of FOSS Applications / Employability LibreOffice – Writer, Calc, Writing letters, documents, creating spreadsheets, tables, Impress making presentations, desktop publishing LibreOffice – Base, Draw, Managing databases, Drawing, doing simple Mathematical Math operations For Commerce IT Students Drupal Drupal is a free and open source content management system (CMS). -
Biovia Materials Studio Visualizer Datasheet
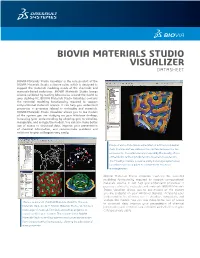
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12 -

Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins
Article Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins Marcus Purse 1, Grace Edmund 1, Stephen Hall 1, Brendan Howlin 1,* , Ian Hamerton 2 and Stephen Till 3 1 Department of Chemistry, Faculty of Engineering and Physical Sciences, University of Surrey, Guilford, Surrey GU2 7XH, UK; [email protected] (M.P.); [email protected] (G.E.); [email protected] (S.H.) 2 Bristol Composites Institute (ACCIS), Department of Aerospace Engineering, School of Civil, Aerospace, and Mechanical Engineering, University of Bristol, Bristol BS8 1TR, UK; [email protected] 3 Defence Science and Technology Laboratory, Porton Down, Salisbury SP4 0JQ, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-1483-686-248 Received: 6 March 2019; Accepted: 23 March 2019; Published: 28 March 2019 Abstract: The thermal decomposition of polyphenolic resins was studied by reactive molecular dynamics (RMD) simulation at elevated temperatures. Atomistic models of the polyphenolic resins to be used in the RMD were constructed using an automatic method which calls routines from the software package Materials Studio. In order to validate the models, simulated densities and heat capacities were compared with experimental values. The most suitable combination of force field and thermostat for this system was the Forcite force field with the Nosé–Hoover thermostat, which gave values of heat capacity closest to those of the experimental values. Simulated densities approached a final density of 1.05–1.08 g/cm3 which compared favorably with the experimental values of 1.16–1.21 g/cm3 for phenol-formaldehyde resins. -

Pymol Modelling Workshop
PyMOL Modelling Workshop My website: http://pldserver1.biochem.queensu.ca/~rlc/work/teaching/BCHM442/ There you will find links to download the latest educational version of PyMOL as well as a link to my “Introduction to PyMOL”, which in turn has links to other people's PyMOL tutorials. Note also the PyMOL Wiki: http://pymolwiki.org. Structure files can be found by searching the Protein Data Bank (PDB) for structure: easy to remember website http://www.pdb.org. There is also the PDBe (PDB Europe, http://www.pdbe.org) that contains the same databank of structures, but with a different web interface for searching for structures and a different set of tools for analyzing structures. What is in a PDB file? Lots of information in the “header” (the section of the file preceding the actual atomic coordinates) as well as the coordinates for the atoms. When assessing a structure, one needs to take account of the resolution and R-factor, error estimates and missing residues. There is information about the sequence that was used to determine the structure with a sequence database reference. There is also information about the biological unit. In the case of crystal structures the biological unit may need to be generated by applying crystallographic symmetry operators, although there are web sites that also try to provide that information. (e.g. http://www.ebi.ac.uk/msd-srv/pisa/). Warning: One cannot blindly trust a crystal structure to be providing you with a completely accurate picture of reality. Reading the paper that describes the structure is a good start! Outline of PyMOL usage PyMOL is a very powerful, scriptable (customizable) tool for making publication-quality figures and performing analyses on structures. -

Dmol Guide to Select a Dmol3 Task 1
DMOL3 GUIDE MATERIALS STUDIO 8.0 Copyright Notice ©2014 Dassault Systèmes. All rights reserved. 3DEXPERIENCE, the Compass icon and the 3DS logo, CATIA, SOLIDWORKS, ENOVIA, DELMIA, SIMULIA, GEOVIA, EXALEAD, 3D VIA, BIOVIA and NETVIBES are commercial trademarks or registered trademarks of Dassault Systèmes or its subsidiaries in the U.S. and/or other countries. All other trademarks are owned by their respective owners. Use of any Dassault Systèmes or its subsidiaries trademarks is subject to their express written approval. Acknowledgments and References To print photographs or files of computational results (figures and/or data) obtained using BIOVIA software, acknowledge the source in an appropriate format. For example: "Computational results obtained using software programs from Dassault Systèmes Biovia Corp.. The ab initio calculations were performed with the DMol3 program, and graphical displays generated with Materials Studio." BIOVIA may grant permission to republish or reprint its copyrighted materials. Requests should be submitted to BIOVIA Support, either through electronic mail to [email protected], or in writing to: BIOVIA Support 5005 Wateridge Vista Drive, San Diego, CA 92121 USA Contents DMol3 1 Setting up a molecular dynamics calculation20 Introduction 1 Choosing an ensemble 21 Further Information 1 Defining the time step 21 Tasks in DMol3 2 Defining the thermostat control 21 Energy 3 Constraints during dynamics 21 Setting up the calculation 3 Setting up a transition state calculation 22 Dynamics 4 Which method to use? -

Atomistic Molecular Dynamics Simulation of Graphene- Isoprene Nanocomposites
Atomistic molecular dynamics simulation of graphene- isoprene nanocomposites P Chanlert1, J Wong-Ekkabut2, W Liewrian3,4,5 and T Sutthibutpong3,4,5* 1 Program of Physics, Faculty of Science and Technology, Songkhla Rajabhat University, Songkhla, 90000, Thailand 2 Department of Physics, Faculty of Science, Kasetsart University, Bangkok 10900, Thailand 3 Theoretical and Computational Physics Group, Department of Physics, Faculty of Science, King Mongkut’s University of Technology Thonburi (KMUTT), Bangkok 10140, Thailand 4 Theoretical and Computational Science Center (TaCS), Science Laboratory Building, Faculty of Science, King Mongkut’s University of Technology Thonburi (KMUTT), Bangkok 10140, Thailand 5 Thailand Center of Excellence in Physics (ThEP Center), Commission on Higher Education, Bangkok 10400, Thailand *email: [email protected] Abstract. A series of atomistic molecular dynamics simulations were performed for 4-mer isoprene molecules confined between graphene sheets with varied graphene separations in order to observe the finite size effects arose from the van der Waals interactions between the isoprene oligomer, forming high density shells at the graphene interfaces. For the small confinement regions, 1.24 nm and 2.21 nm, local density of isoprene at the graphene interface becomes higher than 10 times the bulk density. These results provided the further insights towards the rational design of nanostructures based on graphene sheets and natural rubber polymer. 1. Introduction Composite material is the combination between two materials or more where chemical reaction between these materials is absent. In composites, one with higher quantity is called matrix while those with lesser quantity are called fillers. In general, adding fillers into matrix material enhances both physical and mechanical properties in some ways. -

Molecular Dynamics Simulations of Molecular Diffusion Equilibrium and Breakdown Mechanism of Oil-Impregnated Pressboard with Water Impurity
polymers Article Molecular Dynamics Simulations of Molecular Diffusion Equilibrium and Breakdown Mechanism of Oil-Impregnated Pressboard with Water Impurity Yi Guan 1,2, Ming-He Chi 1,2, Wei-Feng Sun 1,2,*, Qing-Guo Chen 1,2 and Xin-Lao Wei 1,2 1 Heilongjiang Provincial Key Laboratory of Dielectric Engineering, School of Electrical and Electronic Engineering, Harbin University of Science and Technology, Harbin 150080, China; [email protected] (Y.G.); [email protected] (M.-H.C.); [email protected] (Q.-G.C.); [email protected] (X.-L.W.) 2 Key Laboratory of Engineering Dielectrics and Its Application, Ministry of Education, Harbin University of Science and Technology, Harbin 150080, China * Correspondence: [email protected]; Tel.: +86-15846592798 Received: 30 October 2018; Accepted: 15 November 2018; Published: 16 November 2018 Abstract: The water molecule migration and aggregation behaviors in oil-impregnated pressboard are investigated by molecular dynamics simulations in combination with Monte Carlo molecular simulation technique. The free energy and phase diagram of H2O-dodecylbenzene (DDB) and H2O-cellulose mixtures are calculated by Monte Carlo technique combined with the modified Flory-Huggins model, demonstrating that H2O molecules can hardly dissolved with infinitesimal content in cellulose system at temperature lower than 650 K, based on which the oil/cellulose layered structure with water impurity representing three-phase coexistence in oil-impregnated pressboard are modeled and performed for molecular dynamics. The molecular dynamics of H2O/DDB/cellulose three-phase mixture simulating oil-paper insulating system with H2O impurity indicates that DDB molecules can thermally intrude into the cellulose-water interface so as to separate the water phase and cellulose fiber. -

What's New in Biovia Materials Studio 2020
WHAT’S NEW IN BIOVIA MATERIALS STUDIO 2020 Datasheet BIOVIA Materials Studio 2020 is the latest release of BIOVIA’s predictive science tools for chemistry and materials science research. Materials Studio empowers researchers to understand the relationships between a material’s mo- lecular or crystal structure and its properties in order to make more informed decisions about materials research and development. More often than not materials performance is influenced by phenomena at multiple scales. Scientists using Materials Studio 2020 have an extensive suite of world class solvers and parameter sets operating from atoms to microscale for simulating more materials and more properties than ever before. Furthermore, the predicted properties can now be used in multi-physics modeling and in systems modeling software such as SIMULIA Abaqus and CATIA Dymola to predict macroscopic behaviors. In this way multiscale simulations can be used to solve some of the toughest pro- blems in materials design and product optimization. BETTER MATERIALS - BETTER BATTERIES Safe, fast charging batteries with high energy density and long life are urgently needed for a host of applications - not least for the electrification of all modes of transportation as an alternative to fossil fuel energy sources. Battery design relies on a complex interplay between thermal, mechanical and chemical processes from the smallest scales of the material (electronic structure) through to the geometry of the battery cell and pack design. Improvements to the component materials used in batteries and capacitors are fundamental to providing the advances in performance needed. Materials Studio provides new functionality to enable the simula- tion of key materials parameters for both liquid electrolytes and electrode components. -

Enhancing Information Resources and Instruction with Computational and Chemical Software Jeremy R
Purdue University Purdue e-Pubs Libraries Faculty and Staff rP esentations Purdue Libraries 2008 Enhancing information resources and instruction with computational and chemical software Jeremy R. Garritano Purdue University, [email protected] Follow this and additional works at: http://docs.lib.purdue.edu/lib_fspres Part of the Library and Information Science Commons Recommended Citation Garritano, Jeremy R., "Enhancing information resources and instruction with computational and chemical software" (2008). Libraries Faculty and Staff Presentations. Paper 25. http://docs.lib.purdue.edu/lib_fspres/25 This document has been made available through Purdue e-Pubs, a service of the Purdue University Libraries. Please contact [email protected] for additional information. Enhancing Information Resources & Instruction with Computational & Chemical Software Jeremy R. Garritano [email protected] Chemical Information Specialist Assistant Professor of Library Science M. G. Mellon Library of Chemistry Today’s talk • Background • Development and Justification • Resources and Implementation • Marketing • Current Usage • Next Steps Background – The Writing on the Wall • Current Journal Room – Large space – Decreasing number of “browsers” – Push to move online only with print journals began in mid-2006 • Two adjacent rooms – Packed with volumes of Chemical Abstracts Space: Before ← My office Table → Main reading room Current Journal Room Chem. Abs. – Chem. Abs. – Abstracts Abstracts and Indexes Space: Before Solution – Repurpose Space 3 Rooms: • Remove Chemical -
![Trends in Atomistic Simulation Software Usage [1.3]](https://docslib.b-cdn.net/cover/7978/trends-in-atomistic-simulation-software-usage-1-3-1207978.webp)
Trends in Atomistic Simulation Software Usage [1.3]
A LiveCoMS Perpetual Review Trends in atomistic simulation software usage [1.3] Leopold Talirz1,2,3*, Luca M. Ghiringhelli4, Berend Smit1,3 1Laboratory of Molecular Simulation (LSMO), Institut des Sciences et Ingenierie Chimiques, Valais, École Polytechnique Fédérale de Lausanne, CH-1951 Sion, Switzerland; 2Theory and Simulation of Materials (THEOS), Faculté des Sciences et Techniques de l’Ingénieur, École Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland; 3National Centre for Computational Design and Discovery of Novel Materials (MARVEL), École Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland; 4The NOMAD Laboratory at the Fritz Haber Institute of the Max Planck Society and Humboldt University, Berlin, Germany This LiveCoMS document is Abstract Driven by the unprecedented computational power available to scientific research, the maintained online on GitHub at https: use of computers in solid-state physics, chemistry and materials science has been on a continuous //github.com/ltalirz/ rise. This review focuses on the software used for the simulation of matter at the atomic scale. We livecoms-atomistic-software; provide a comprehensive overview of major codes in the field, and analyze how citations to these to provide feedback, suggestions, or help codes in the academic literature have evolved since 2010. An interactive version of the underlying improve it, please visit the data set is available at https://atomistic.software. GitHub repository and participate via the issue tracker. This version dated August *For correspondence: 30, 2021 [email protected] (LT) 1 Introduction Gaussian [2], were already released in the 1970s, followed Scientists today have unprecedented access to computa- by force-field codes, such as GROMOS [3], and periodic tional power.