Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

View of HER2: Human Epidermal Growth Factor Receptor 2; TNBC: Triple-Negative Breast Resistance to Systemic Therapy in Patients with Breast Cancer
Wen et al. Cancer Cell Int (2018) 18:128 https://doi.org/10.1186/s12935-018-0625-9 Cancer Cell International PRIMARY RESEARCH Open Access Sulbactam‑enhanced cytotoxicity of doxorubicin in breast cancer cells Shao‑hsuan Wen1†, Shey‑chiang Su2†, Bo‑huang Liou3, Cheng‑hao Lin1 and Kuan‑rong Lee1* Abstract Background: Multidrug resistance (MDR) is a major obstacle in breast cancer treatment. The predominant mecha‑ nism underlying MDR is an increase in the activity of adenosine triphosphate (ATP)-dependent drug efux trans‑ porters. Sulbactam, a β-lactamase inhibitor, is generally combined with β-lactam antibiotics for treating bacterial infections. However, sulbactam alone can be used to treat Acinetobacter baumannii infections because it inhibits the expression of ATP-binding cassette (ABC) transporter proteins. This is the frst study to report the efects of sulbactam on mammalian cells. Methods: We used the breast cancer cell lines as a model system to determine whether sulbactam afects cancer cells. The cell viabilities in the present of doxorubicin with or without sulbactam were measured by MTT assay. Protein identities and the changes in protein expression levels in the cells after sulbactam and doxorubicin treatment were determined using LC–MS/MS. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) was used to analyze the change in mRNA expression levels of ABC transporters after treatment of doxorubicin with or without sulbactam. The efux of doxorubicin was measures by the doxorubicin efux assay. Results: MTT assay revealed that sulbactam enhanced the cytotoxicity of doxorubicin in breast cancer cells. The results of proteomics showed that ABC transporter proteins and proteins associated with the process of transcription and initiation of translation were reduced. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -
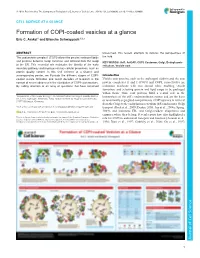
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -

Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability
The Journal of Neuroscience, March 7, 2018 • 38(10):2615–2630 • 2615 Neurobiology of Disease Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability Emin Kuliyev,1 Sebastien Gingras,4 XClifford S. Guy,1 Sherie Howell,2 Peter Vogel,3 and XStephane Pelletier1 1Departments of Immunology, 2Pathology, 3Veterinary Pathology Core, Advanced Histology Core, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105, and 4Department of Immunology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15213 Members of the SCY1-like (SCYL) family of protein kinases are evolutionarily conserved and ubiquitously expressed proteins character- ized by an N-terminal pseudokinase domain, centrally located Huntingtin, elongation factor 3, protein phosphatase 2A, yeast kinase TOR1 repeats, and an overall disorganized C-terminal segment. In mammals, three family members encoded by genes Scyl1, Scyl2, and Scyl3 have been described. Studies have pointed to a role for SCYL1 and SCYL2 in regulating neuronal function and viability in mice and humans, but little is known about the biological function of SCYL3. Here, we show that the biochemical and cell biological properties of SCYL3aresimilartothoseofSCYL1andbothproteinsworkinconjunctiontomaintainmotorneuronviability.Specifically,althoughlack of Scyl3 in mice has no apparent effect on embryogenesis and postnatal life, it accelerates the onset of the motor neuron disorder caused by Scyl1 deficiency. Growth abnormalities, motor dysfunction, hindlimb paralysis, muscle wasting, neurogenic atrophy, motor neuron degeneration, and loss of large-caliber axons in peripheral nerves occurred at an earlier age in Scyl1/Scyl3 double-deficient mice than in Scyl1-deficientmice.DiseaseonsetalsocorrelatedwiththemislocalizationofTDP-43inspinalmotorneurons,suggestingthatSCYL1and SCYL3 regulate TDP-43 proteostasis. Together, our results demonstrate an overlapping role for SCYL1 and SCYL3 in vivo and highlight the importance the SCYL family of proteins in regulating neuronal function and survival. -

Dissertation
DISSERTATION submitted to the Combined Faculty of Natural Sciences and Mathematics of the Ruperto Carola University Heidelberg, Germany for the degree of Doctor of Natural Sciences Presented by Manu Jain Goyal (M.Sc, Biotechnology) born in Kota, India Oral examination: 03.07.2019 Paralog specific role of COPI pathway in P19 neuronal differentiation Referees: Prof. Dr. Felix Wieland Dr. Julien Béthune This work has been carried out at the Biochemistry Center of the University of Heidelberg from January 2015 to January 2019 under the supervision of Dr. Julien Béthune. Acknowledgement I would like to thank Julien for giving me opportunity to be part of his team, for supervising me throughout my PhD. I appreciate that all the scientific discussions we have had helped me to improve my knowledge and thinking scientifically. Thank you so much for giving freedom in the lab to work independently and for handling the situations in a best possible way. Next, I would like to thank my TAC members Prof. Dr. Felix and Prof. Dr. Walter for giving suggestions and inputs for my project. Also providing me antibodies and equipments for my experiments. I would like to thank my examiners Prof Dr. Thomas Söllner and Prof. Dr. Marius Lemberg for considering it. I would like to thank all my team members and special thanks to Petra, Cinthia and Isabel for always supporting and helping me. Additionally, I would like to thank Alice and Laura for socializing together outside the BZH so that I can survive far away from my home country. I admire all the moments I had with you all. -

Consequences of Mitotic Loss of Heterozygosity on Genomic Imprinting in Mouse Embryonic Stem Cells
CONSEQUENCES OF MITOTIC LOSS OF HETEROZYGOSITY ON GENOMIC IMPRINTING IN MOUSE EMBRYONIC STEM CELLS by RACHEL LEIGH ELVES B.Sc., University of British Columbia, 2004 A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIRMENTS FOR THE DEGREE OF MASTER OF SCIENCE in THE FACULTY OF GRADUATE STUDIES (Medical Genetics) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) August 2008 © Rachel Leigh Elves, 2008 Abstract Epigenetic differences between maternally inherited and paternally inherited chromosomes, such as CpG methylation, render the maternal and paternal genome functionally inequivalent, a phenomenon called genomic imprinting. This functional inequivalence is exemplified with imprinted genes, whose expression is parent-of-origin specific. The dosage of imprinted gene expression is disrupted in cells with uniparental disomy (UPD), which is an unequal parental contribution to the genome. I have derived mouse embryonic stem (ES) cell sub-lines with maternal UPD (mUPD) for mouse chromosome 6 (MMU6) to characterize regulation and maintenance of imprinted gene expression. The main finding from this study is that maintenance of imprinting in mitotic UPD is extremely variable. Imprint maintenance was shown to vary from gene to gene, and to vary between ES cell lines depending on the mechanism of loss of heterozygosity (LOH) in that cell line. Certain genes analyzed, such as Peg10 , Sgce , Peg1 , and Mit1 showed abnormal expression in ES cell lines for which they were mUPD. These abnormal expression levels are similar to that observed in ES cells with meiotically-derived full genome mUPD (parthenogenetic ES cells). Imprinted CpG methylation at the Peg1 promoter was found to be abnormal in all sub-lines with mUPD for Peg1 . -

Supplementary Table S2
1-high in cerebrotropic Gene P-value patients Definition BCHE 2.00E-04 1 Butyrylcholinesterase PLCB2 2.00E-04 -1 Phospholipase C, beta 2 SF3B1 2.00E-04 -1 Splicing factor 3b, subunit 1 BCHE 0.00022 1 Butyrylcholinesterase ZNF721 0.00028 -1 Zinc finger protein 721 GNAI1 0.00044 1 Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 GNAI1 0.00049 1 Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 PDE1B 0.00069 -1 Phosphodiesterase 1B, calmodulin-dependent MCOLN2 0.00085 -1 Mucolipin 2 PGCP 0.00116 1 Plasma glutamate carboxypeptidase TMX4 0.00116 1 Thioredoxin-related transmembrane protein 4 C10orf11 0.00142 1 Chromosome 10 open reading frame 11 TRIM14 0.00156 -1 Tripartite motif-containing 14 APOBEC3D 0.00173 -1 Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D ANXA6 0.00185 -1 Annexin A6 NOS3 0.00209 -1 Nitric oxide synthase 3 SELI 0.00209 -1 Selenoprotein I NYNRIN 0.0023 -1 NYN domain and retroviral integrase containing ANKFY1 0.00253 -1 Ankyrin repeat and FYVE domain containing 1 APOBEC3F 0.00278 -1 Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F EBI2 0.00278 -1 Epstein-Barr virus induced gene 2 ETHE1 0.00278 1 Ethylmalonic encephalopathy 1 PDE7A 0.00278 -1 Phosphodiesterase 7A HLA-DOA 0.00305 -1 Major histocompatibility complex, class II, DO alpha SOX13 0.00305 1 SRY (sex determining region Y)-box 13 ABHD2 3.34E-03 1 Abhydrolase domain containing 2 MOCS2 0.00334 1 Molybdenum cofactor synthesis 2 TTLL6 0.00365 -1 Tubulin tyrosine ligase-like family, member 6 SHANK3 0.00394 -1 SH3 and multiple ankyrin repeat domains 3 ADCY4 0.004 -1 Adenylate cyclase 4 CD3D 0.004 -1 CD3d molecule, delta (CD3-TCR complex) (CD3D), transcript variant 1, mRNA. -

The Role of ARF Family Proteins and Their Regulators and Effectors in Cancer Progression: a Therapeutic Perspective
fcell-08-00217 April 17, 2020 Time: 19:19 # 1 REVIEW published: 21 April 2020 doi: 10.3389/fcell.2020.00217 The Role of ARF Family Proteins and Their Regulators and Effectors in Cancer Progression: A Therapeutic Perspective Cristina Casalou†, Andreia Ferreira† and Duarte C. Barral* CEDOC, NOVA Medical School, Faculdade de Ciências Médicas, Universidade NOVA de Lisboa, Lisbon, Portugal The Adenosine diphosphate-Ribosylation Factor (ARF) family belongs to the RAS superfamily of small GTPases and is involved in a wide variety of physiological processes, such as cell proliferation, motility and differentiation by regulating membrane Edited by: traffic and associating with the cytoskeleton. Like other members of the RAS Sunil Kumar Verma, superfamily, ARF family proteins are activated by Guanine nucleotide Exchange Factors Centre for Cellular & Molecular (GEFs) and inactivated by GTPase-Activating Proteins (GAPs). When active, they bind Biology (CCMB), India effectors, which mediate downstream functions. Several studies have reported that Reviewed by: Wei-Hsiung Yang, cancer cells are able to subvert membrane traffic regulators to enhance migration and Mercer University, United States invasion. Indeed, members of the ARF family, including ARF-Like (ARL) proteins have Ira Daar, National Cancer Institute (NCI), been implicated in tumorigenesis and progression of several types of cancer. Here, we United States review the role of ARF family members, their GEFs/GAPs and effectors in tumorigenesis *Correspondence: and cancer progression, highlighting the ones that can have a pro-oncogenic behavior Duarte C. Barral or function as tumor suppressors. Moreover, we propose possible mechanisms and [email protected] approaches to target these proteins, toward the development of novel therapeutic †These authors have contributed equally to this work strategies to impair tumor progression.