A Hierarchical Voxel Hash for Fast 3D Nearest Neighbor Lookup
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

4 Hash Tables and Associative Arrays
4 FREE Hash Tables and Associative Arrays If you want to get a book from the central library of the University of Karlsruhe, you have to order the book in advance. The library personnel fetch the book from the stacks and deliver it to a room with 100 shelves. You find your book on a shelf numbered with the last two digits of your library card. Why the last digits and not the leading digits? Probably because this distributes the books more evenly among the shelves. The library cards are numbered consecutively as students sign up, and the University of Karlsruhe was founded in 1825. Therefore, the students enrolled at the same time are likely to have the same leading digits in their card number, and only a few shelves would be in use if the leadingCOPY digits were used. The subject of this chapter is the robust and efficient implementation of the above “delivery shelf data structure”. In computer science, this data structure is known as a hash1 table. Hash tables are one implementation of associative arrays, or dictio- naries. The other implementation is the tree data structures which we shall study in Chap. 7. An associative array is an array with a potentially infinite or at least very large index set, out of which only a small number of indices are actually in use. For example, the potential indices may be all strings, and the indices in use may be all identifiers used in a particular C++ program.Or the potential indices may be all ways of placing chess pieces on a chess board, and the indices in use may be the place- ments required in the analysis of a particular game. -

Two Algorithms for Nearest-Neighbor Search in High Dimensions
Two Algorithms for Nearest-Neighbor Search in High Dimensions Jon M. Kleinberg∗ February 7, 1997 Abstract Representing data as points in a high-dimensional space, so as to use geometric methods for indexing, is an algorithmic technique with a wide array of uses. It is central to a number of areas such as information retrieval, pattern recognition, and statistical data analysis; many of the problems arising in these applications can involve several hundred or several thousand dimensions. We consider the nearest-neighbor problem for d-dimensional Euclidean space: we wish to pre-process a database of n points so that given a query point, one can ef- ficiently determine its nearest neighbors in the database. There is a large literature on algorithms for this problem, in both the exact and approximate cases. The more sophisticated algorithms typically achieve a query time that is logarithmic in n at the expense of an exponential dependence on the dimension d; indeed, even the average- case analysis of heuristics such as k-d trees reveals an exponential dependence on d in the query time. In this work, we develop a new approach to the nearest-neighbor problem, based on a method for combining randomly chosen one-dimensional projections of the underlying point set. From this, we obtain the following two results. (i) An algorithm for finding ε-approximate nearest neighbors with a query time of O((d log2 d)(d + log n)). (ii) An ε-approximate nearest-neighbor algorithm with near-linear storage and a query time that improves asymptotically on linear search in all dimensions. -

Rank-Approximate Nearest Neighbor Search: Retaining Meaning and Speed in High Dimensions
Rank-Approximate Nearest Neighbor Search: Retaining Meaning and Speed in High Dimensions Parikshit Ram, Dongryeol Lee, Hua Ouyang and Alexander G. Gray Computational Science and Engineering, Georgia Institute of Technology Atlanta, GA 30332 p.ram@,dongryel@cc.,houyang@,agray@cc. gatech.edu { } Abstract The long-standing problem of efficient nearest-neighbor (NN) search has ubiqui- tous applications ranging from astrophysics to MP3 fingerprinting to bioinformat- ics to movie recommendations. As the dimensionality of the dataset increases, ex- act NN search becomes computationally prohibitive; (1+) distance-approximate NN search can provide large speedups but risks losing the meaning of NN search present in the ranks (ordering) of the distances. This paper presents a simple, practical algorithm allowing the user to, for the first time, directly control the true accuracy of NN search (in terms of ranks) while still achieving the large speedups over exact NN. Experiments on high-dimensional datasets show that our algorithm often achieves faster and more accurate results than the best-known distance-approximate method, with much more stable behavior. 1 Introduction In this paper, we address the problem of nearest-neighbor (NN) search in large datasets of high dimensionality. It is used for classification (k-NN classifier [1]), categorizing a test point on the ba- sis of the classes in its close neighborhood. Non-parametric density estimation uses NN algorithms when the bandwidth at any point depends on the ktℎ NN distance (NN kernel density estimation [2]). NN algorithms are present in and often the main cost of most non-linear dimensionality reduction techniques (manifold learning [3, 4]) to obtain the neighborhood of every point which is then pre- served during the dimension reduction. -

Prefix Hash Tree an Indexing Data Structure Over Distributed Hash
Prefix Hash Tree An Indexing Data Structure over Distributed Hash Tables Sriram Ramabhadran ∗ Sylvia Ratnasamy University of California, San Diego Intel Research, Berkeley Joseph M. Hellerstein Scott Shenker University of California, Berkeley International Comp. Science Institute, Berkeley and and Intel Research, Berkeley University of California, Berkeley ABSTRACT this lookup interface has allowed a wide variety of Distributed Hash Tables are scalable, robust, and system to be built on top DHTs, including file sys- self-organizing peer-to-peer systems that support tems [9, 27], indirection services [30], event notifi- exact match lookups. This paper describes the de- cation [6], content distribution networks [10] and sign and implementation of a Prefix Hash Tree - many others. a distributed data structure that enables more so- phisticated queries over a DHT. The Prefix Hash DHTs were designed in the Internet style: scala- Tree uses the lookup interface of a DHT to con- bility and ease of deployment triumph over strict struct a trie-based structure that is both efficient semantics. In particular, DHTs are self-organizing, (updates are doubly logarithmic in the size of the requiring no centralized authority or manual con- domain being indexed), and resilient (the failure figuration. They are robust against node failures of any given node in the Prefix Hash Tree does and easily accommodate new nodes. Most impor- not affect the availability of data stored at other tantly, they are scalable in the sense that both la- nodes). tency (in terms of the number of hops per lookup) and the local state required typically grow loga- Categories and Subject Descriptors rithmically in the number of nodes; this is crucial since many of the envisioned scenarios for DHTs C.2.4 [Comp. -

Hash Tables & Searching Algorithms
Search Algorithms and Tables Chapter 11 Tables • A table, or dictionary, is an abstract data type whose data items are stored and retrieved according to a key value. • The items are called records. • Each record can have a number of data fields. • The data is ordered based on one of the fields, named the key field. • The record we are searching for has a key value that is called the target. • The table may be implemented using a variety of data structures: array, tree, heap, etc. Sequential Search public static int search(int[] a, int target) { int i = 0; boolean found = false; while ((i < a.length) && ! found) { if (a[i] == target) found = true; else i++; } if (found) return i; else return –1; } Sequential Search on Tables public static int search(someClass[] a, int target) { int i = 0; boolean found = false; while ((i < a.length) && !found){ if (a[i].getKey() == target) found = true; else i++; } if (found) return i; else return –1; } Sequential Search on N elements • Best Case Number of comparisons: 1 = O(1) • Average Case Number of comparisons: (1 + 2 + ... + N)/N = (N+1)/2 = O(N) • Worst Case Number of comparisons: N = O(N) Binary Search • Can be applied to any random-access data structure where the data elements are sorted. • Additional parameters: first – index of the first element to examine size – number of elements to search starting from the first element above Binary Search • Precondition: If size > 0, then the data structure must have size elements starting with the element denoted as the first element. In addition, these elements are sorted. -

Nearest Neighbor Search in Google Correlate
Nearest Neighbor Search in Google Correlate Dan Vanderkam Robert Schonberger Henry Rowley Google Inc Google Inc Google Inc 76 9th Avenue 76 9th Avenue 1600 Amphitheatre Parkway New York, New York 10011 New York, New York 10011 Mountain View, California USA USA 94043 USA [email protected] [email protected] [email protected] Sanjiv Kumar Google Inc 76 9th Avenue New York, New York 10011 USA [email protected] ABSTRACT queries are then noted, and a model that estimates influenza This paper presents the algorithms which power Google Cor- incidence based on present query data is created. relate[8], a tool which finds web search terms whose popu- This estimate is valuable because the most recent tradi- larity over time best matches a user-provided time series. tional estimate of the ILI rate is only available after a two Correlate was developed to generalize the query-based mod- week delay. Indeed, there are many fields where estimating eling techniques pioneered by Google Flu Trends and make the present[1] is important. Other health issues and indica- them available to end users. tors in economics, such as unemployment, can also benefit Correlate searches across millions of candidate query time from estimates produced using the techniques developed for series to find the best matches, returning results in less than Google Flu Trends. 200 milliseconds. Its feature set and requirements present Google Flu Trends relies on a multi-hour batch process unique challenges for Approximate Nearest Neighbor (ANN) to find queries that correlate to the ILI time series. Google search techniques. In this paper, we present Asymmetric Correlate allows users to create their own versions of Flu Hashing (AH), the technique used by Correlate, and show Trends in real time. -

Introduction to Hash Table and Hash Function
Introduction to Hash Table and Hash Function This is a short introduction to Hashing mechanism Introduction Is it possible to design a search of O(1)– that is, one that has a constant search time, no matter where the element is located in the list? Let’s look at an example. We have a list of employees of a fairly small company. Each of 100 employees has an ID number in the range 0 – 99. If we store the elements (employee records) in the array, then each employee’s ID number will be an index to the array element where this employee’s record will be stored. In this case once we know the ID number of the employee, we can directly access his record through the array index. There is a one-to-one correspondence between the element’s key and the array index. However, in practice, this perfect relationship is not easy to establish or maintain. For example: the same company might use employee’s five-digit ID number as the primary key. In this case, key values run from 00000 to 99999. If we want to use the same technique as above, we need to set up an array of size 100,000, of which only 100 elements will be used: Obviously it is very impractical to waste that much storage. But what if we keep the array size down to the size that we will actually be using (100 elements) and use just the last two digits of key to identify each employee? For example, the employee with the key number 54876 will be stored in the element of the array with index 76. -

Balanced Search Trees and Hashing Balanced Search Trees
Advanced Implementations of Tables: Balanced Search Trees and Hashing Balanced Search Trees Binary search tree operations such as insert, delete, retrieve, etc. depend on the length of the path to the desired node The path length can vary from log2(n+1) to O(n) depending on how balanced or unbalanced the tree is The shape of the tree is determined by the values of the items and the order in which they were inserted CMPS 12B, UC Santa Cruz Binary searching & introduction to trees 2 Examples 10 40 20 20 60 30 40 10 30 50 70 50 Can you get the same tree with 60 different insertion orders? 70 CMPS 12B, UC Santa Cruz Binary searching & introduction to trees 3 2-3 Trees Each internal node has two or three children All leaves are at the same level 2 children = 2-node, 3 children = 3-node CMPS 12B, UC Santa Cruz Binary searching & introduction to trees 4 2-3 Trees (continued) 2-3 trees are not binary trees (duh) A 2-3 tree of height h always has at least 2h-1 nodes i.e. greater than or equal to a binary tree of height h A 2-3 tree with n nodes has height less than or equal to log2(n+1) i.e less than or equal to the height of a full binary tree with n nodes CMPS 12B, UC Santa Cruz Binary searching & introduction to trees 5 Definition of 2-3 Trees T is a 2-3 tree of height h if T is empty (height 0), OR T is of the form r TL TR Where r is a node that contains one data item and TL and TR are 2-3 trees, each of height h-1, and the search key of r is greater than any in TL and less than any in TR, OR CMPS 12B, UC Santa Cruz Binary searching & introduction to trees 6 Definition of 2-3 Trees (continued) T is of the form r TL TM TR Where r is a node that contains two data items and TL, TM, and TR are 2-3 trees, each of height h-1, and the smaller search key of r is greater than any in TL and less than any in TM and the larger search key in r is greater than any in TM and smaller than any in TR. -

R-Trees with Voronoi Diagrams for Efficient Processing of Spatial
VoR-Tree: R-trees with Voronoi Diagrams for Efficient Processing of Spatial Nearest Neighbor Queries∗ y Mehdi Sharifzadeh Cyrus Shahabi Google University of Southern California [email protected] [email protected] ABSTRACT A very important class of spatial queries consists of nearest- An important class of queries, especially in the geospatial neighbor (NN) query and its variations. Many studies in domain, is the class of nearest neighbor (NN) queries. These the past decade utilize R-trees as their underlying index queries search for data objects that minimize a distance- structures to address NN queries efficiently. The general ap- based function with reference to one or more query objects. proach is to use R-tree in two phases. First, R-tree's hierar- Examples are k Nearest Neighbor (kNN) [13, 5, 7], Reverse chical structure is used to quickly arrive to the neighborhood k Nearest Neighbor (RkNN) [8, 15, 16], k Aggregate Nearest of the result set. Second, the R-tree nodes intersecting with Neighbor (kANN) [12] and skyline queries [1, 11, 14]. The the local neighborhood (Search Region) of an initial answer applications of NN queries are numerous in geospatial deci- are investigated to find all the members of the result set. sion making, location-based services, and sensor networks. While R-trees are very efficient for the first phase, they usu- The introduction of R-trees [3] (and their extensions) for ally result in the unnecessary investigation of many nodes indexing multi-dimensional data marked a new era in de- that none or only a small subset of their including points veloping novel R-tree-based algorithms for various forms belongs to the actual result set. -

Dimensional Testing for Reverse K-Nearest Neighbor Search
University of Southern Denmark Dimensional testing for reverse k-nearest neighbor search Casanova, Guillaume; Englmeier, Elias; Houle, Michael E.; Kröger, Peer; Nett, Michael; Schubert, Erich; Zimek, Arthur Published in: Proceedings of the VLDB Endowment DOI: 10.14778/3067421.3067426 Publication date: 2017 Document version: Final published version Document license: CC BY-NC-ND Citation for pulished version (APA): Casanova, G., Englmeier, E., Houle, M. E., Kröger, P., Nett, M., Schubert, E., & Zimek, A. (2017). Dimensional testing for reverse k-nearest neighbor search. Proceedings of the VLDB Endowment, 10(7), 769-780. https://doi.org/10.14778/3067421.3067426 Go to publication entry in University of Southern Denmark's Research Portal Terms of use This work is brought to you by the University of Southern Denmark. Unless otherwise specified it has been shared according to the terms for self-archiving. If no other license is stated, these terms apply: • You may download this work for personal use only. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying this open access version If you believe that this document breaches copyright please contact us providing details and we will investigate your claim. Please direct all enquiries to [email protected] Download date: 05. Oct. 2021 Dimensional Testing for Reverse k-Nearest Neighbor Search Guillaume Casanova Elias Englmeier Michael E. Houle ONERA-DCSD, France LMU Munich, Germany NII, Tokyo, -

Hash Tables Hash Tables a "Faster" Implementation for Map Adts
Hash Tables Hash Tables A "faster" implementation for Map ADTs Outline What is hash function? translation of a string key into an integer Consider a few strategies for implementing a hash table linear probing quadratic probing separate chaining hashing Big Ohs using different data structures for a Map ADT? Data Structure put get remove Unsorted Array Sorted Array Unsorted Linked List Sorted Linked List Binary Search Tree A BST was used in OrderedMap<K,V> Hash Tables Hash table: another data structure Provides virtually direct access to objects based on a key (a unique String or Integer) key could be your SID, your telephone number, social security number, account number, … Must have unique keys Each key is associated with–mapped to–a value Hashing Must convert keys such as "555-1234" into an integer index from 0 to some reasonable size Elements can be found, inserted, and removed using the integer index as an array index Insert (called put), find (get), and remove must use the same "address calculator" which we call the Hash function Hashing Can make String or Integer keys into integer indexes by "hashing" Need to take hashCode % array size Turn “S12345678” into an int 0..students.length Ideally, every key has a unique hash value Then the hash value could be used as an array index, however, Ideal is impossible Some keys will "hash" to the same integer index Known as a collision Need a way to handle collisions! "abc" may hash to the same integer array index as "cba" Hash Tables: Runtime Efficient Lookup time does not grow when n increases A hash table supports fast insertion O(1) fast retrieval O(1) fast removal O(1) Could use String keys each ASCII character equals some unique integer "able" = 97 + 98 + 108 + 101 == 404 Hash method works something like… Convert a String key into an integer that will be in the range of 0 through the maximum capacity-1 Assume the array capacity is 9997 hash(key) AAAAAAAA 8482 zzzzzzzz 1273 hash(key) A string of 8 chars Range: 0 .. -
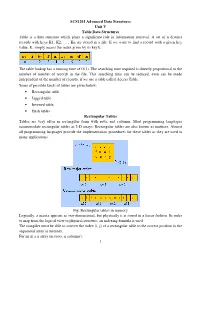
SCS1201 Advanced Data Structures Unit V Table Data Structures Table Is a Data Structure Which Plays a Significant Role in Information Retrieval
SCS1201 Advanced Data Structures Unit V Table Data Structures Table is a data structure which plays a significant role in information retrieval. A set of n distinct records with keys K1, K2, …., Kn are stored in a file. If we want to find a record with a given key value, K, simply access the index given by its key k. The table lookup has a running time of O(1). The searching time required is directly proportional to the number of number of records in the file. This searching time can be reduced, even can be made independent of the number of records, if we use a table called Access Table. Some of possible kinds of tables are given below: • Rectangular table • Jagged table • Inverted table. • Hash tables Rectangular Tables Tables are very often in rectangular form with rows and columns. Most programming languages accommodate rectangular tables as 2-D arrays. Rectangular tables are also known as matrices. Almost all programming languages provide the implementation procedures for these tables as they are used in many applications. Fig. Rectangular tables in memory Logically, a matrix appears as two-dimensional, but physically it is stored in a linear fashion. In order to map from the logical view to physical structure, an indexing formula is used. The compiler must be able to convert the index (i, j) of a rectangular table to the correct position in the sequential array in memory. For an m x n array (m rows, n columns): 1 Each row is indexed from 0 to m-1 Each column is indexed from 0 to n – 1 Item at (i, j) is at sequential position i * n + j Row major order: Assume that the base address is the first location of the memory, so the th Address a ij =storing all the elements in the first(i-1) rows + the number of elements in the i th row up to the j th coloumn = (i-1)*n+j Column major order: Address of a ij = storing all the elements in the first(j-1)th column + The number of elements in the j th column up to the i th rows.