Lecture 2: Simple Classifiers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Descriptive Statistics (Part 2): Interpreting Study Results
Statistical Notes II Descriptive statistics (Part 2): Interpreting study results A Cook and A Sheikh he previous paper in this series looked at ‘baseline’. Investigations of treatment effects can be descriptive statistics, showing how to use and made in similar fashion by comparisons of disease T interpret fundamental measures such as the probability in treated and untreated patients. mean and standard deviation. Here we continue with descriptive statistics, looking at measures more The relative risk (RR), also sometimes known as specific to medical research. We start by defining the risk ratio, compares the risk of exposed and risk and odds, the two basic measures of disease unexposed subjects, while the odds ratio (OR) probability. Then we show how the effect of a disease compares odds. A relative risk or odds ratio greater risk factor, or a treatment, can be measured using the than one indicates an exposure to be harmful, while relative risk or the odds ratio. Finally we discuss the a value less than one indicates a protective effect. ‘number needed to treat’, a measure derived from the RR = 1.2 means exposed people are 20% more likely relative risk, which has gained popularity because of to be diseased, RR = 1.4 means 40% more likely. its clinical usefulness. Examples from the literature OR = 1.2 means that the odds of disease is 20% higher are used to illustrate important concepts. in exposed people. RISK AND ODDS Among workers at factory two (‘exposed’ workers) The probability of an individual becoming diseased the risk is 13 / 116 = 0.11, compared to an ‘unexposed’ is the risk. -

Teacher Guide 12.1 Gambling Page 1
TEACHER GUIDE 12.1 GAMBLING PAGE 1 Standard 12: The student will explain and evaluate the financial impact and consequences of gambling. Risky Business Priority Academic Student Skills Personal Financial Literacy Objective 12.1: Analyze the probabilities involved in winning at games of chance. Objective 12.2: Evaluate costs and benefits of gambling to Simone, Paula, and individuals and society (e.g., family budget; addictive behaviors; Randy meet in the library and the local and state economy). every afternoon to work on their homework. Here is what is going on. Ryan is flipping a coin, and he is not cheating. He has just flipped seven heads in a row. Is Ryan’s next flip more likely to be: Heads; Tails; or Heads and tails are equally likely. Paula says heads because the first seven were heads, so the next one will probably be heads too. Randy says tails. The first seven were heads, so the next one must be tails. Lesson Objectives Simone says that it could be either heads or tails Recognize gambling as a form of risk. because both are equally Calculate the probabilities of winning in games of chance. likely. Who is correct? Explore the potential benefits of gambling for society. Explore the potential costs of gambling for society. Evaluate the personal costs and benefits of gambling. © 2008. Oklahoma State Department of Education. All rights reserved. Teacher Guide 12.1 2 Personal Financial Literacy Vocabulary Dependent event: The outcome of one event affects the outcome of another, changing the TEACHING TIP probability of the second event. This lesson is based on risk. -

Small-Sample Properties of Tests for Heteroscedasticity in the Conditional Logit Model
HEDG Working Paper 06/04 Small-sample properties of tests for heteroscedasticity in the conditional logit model Arne Risa Hole May 2006 ISSN 1751-1976 york.ac.uk/res/herc/hedgwp Small-sample properties of tests for heteroscedasticity in the conditional logit model Arne Risa Hole National Primary Care Research and Development Centre Centre for Health Economics University of York May 16, 2006 Abstract This paper compares the small-sample properties of several asymp- totically equivalent tests for heteroscedasticity in the conditional logit model. While no test outperforms the others in all of the experiments conducted, the likelihood ratio test and a particular variety of the Wald test are found to have good properties in moderate samples as well as being relatively powerful. Keywords: conditional logit, heteroscedasticity JEL classi…cation: C25 National Primary Care Research and Development Centre, Centre for Health Eco- nomics, Alcuin ’A’Block, University of York, York YO10 5DD, UK. Tel.: +44 1904 321404; fax: +44 1904 321402. E-mail address: [email protected]. NPCRDC receives funding from the Department of Health. The views expressed are not necessarily those of the funders. 1 1 Introduction In most applications of the conditional logit model the error term is assumed to be homoscedastic. Recently, however, there has been a growing interest in testing the homoscedasticity assumption in applied work (Hensher et al., 1999; DeShazo and Fermo, 2002). This is partly due to the well-known result that heteroscedasticity causes the coe¢ cient estimates in discrete choice models to be inconsistent (Yatchew and Griliches, 1985), but also re‡ects a behavioural interest in factors in‡uencing the variance of the latent variables in the model (Louviere, 2001; Louviere et al., 2002). -

Odds: Gambling, Law and Strategy in the European Union Anastasios Kaburakis* and Ryan M Rodenberg†
63 Odds: Gambling, Law and Strategy in the European Union Anastasios Kaburakis* and Ryan M Rodenberg† Contemporary business law contributions argue that legal knowledge or ‘legal astuteness’ can lead to a sustainable competitive advantage.1 Past theses and treatises have led more academic research to endeavour the confluence between law and strategy.2 European scholars have engaged in the Proactive Law Movement, recently adopted and incorporated into policy by the European Commission.3 As such, the ‘many futures of legal strategy’ provide * Dr Anastasios Kaburakis is an assistant professor at the John Cook School of Business, Saint Louis University, teaching courses in strategic management, sports business and international comparative gaming law. He holds MS and PhD degrees from Indiana University-Bloomington and a law degree from Aristotle University in Thessaloniki, Greece. Prior to academia, he practised law in Greece and coached basketball at the professional club and national team level. He currently consults international sport federations, as well as gaming operators on regulatory matters and policy compliance strategies. † Dr Ryan Rodenberg is an assistant professor at Florida State University where he teaches sports law analytics. He earned his JD from the University of Washington-Seattle and PhD from Indiana University-Bloomington. Prior to academia, he worked at Octagon, Nike and the ATP World Tour. 1 See C Bagley, ‘What’s Law Got to Do With It?: Integrating Law and Strategy’ (2010) 47 American Business Law Journal 587; C -

Multinomial Logistic Regression
26-Osborne (Best)-45409.qxd 10/9/2007 5:52 PM Page 390 26 MULTINOMIAL LOGISTIC REGRESSION CAROLYN J. ANDERSON LESLIE RUTKOWSKI hapter 24 presented logistic regression Many of the concepts used in binary logistic models for dichotomous response vari- regression, such as the interpretation of parame- C ables; however, many discrete response ters in terms of odds ratios and modeling prob- variables have three or more categories (e.g., abilities, carry over to multicategory logistic political view, candidate voted for in an elec- regression models; however, two major modifica- tion, preferred mode of transportation, or tions are needed to deal with multiple categories response options on survey items). Multicate- of the response variable. One difference is that gory response variables are found in a wide with three or more levels of the response variable, range of experiments and studies in a variety of there are multiple ways to dichotomize the different fields. A detailed example presented in response variable. If J equals the number of cate- this chapter uses data from 600 students from gories of the response variable, then J(J – 1)/2 dif- the High School and Beyond study (Tatsuoka & ferent ways exist to dichotomize the categories. Lohnes, 1988) to look at the differences among In the High School and Beyond study, the three high school students who attended academic, program types can be dichotomized into pairs general, or vocational programs. The students’ of programs (i.e., academic and general, voca- socioeconomic status (ordinal), achievement tional and general, and academic and vocational). test scores (numerical), and type of school How the response variable is dichotomized (nominal) are all examined as possible explana- depends, in part, on the nature of the variable. -

Logit and Ordered Logit Regression (Ver
Getting Started in Logit and Ordered Logit Regression (ver. 3.1 beta) Oscar Torres-Reyna Data Consultant [email protected] http://dss.princeton.edu/training/ PU/DSS/OTR Logit model • Use logit models whenever your dependent variable is binary (also called dummy) which takes values 0 or 1. • Logit regression is a nonlinear regression model that forces the output (predicted values) to be either 0 or 1. • Logit models estimate the probability of your dependent variable to be 1 (Y=1). This is the probability that some event happens. PU/DSS/OTR Logit odelm From Stock & Watson, key concept 9.3. The logit model is: Pr(YXXXFXX 1 | 1= , 2 ,...=k β ) +0 β ( 1 +2 β 1 +βKKX 2 + ... ) 1 Pr(YXXX 1= | 1 , 2k = ,... ) 1−+(eβ0 + βXX 1 1 + β 2 2 + ...βKKX + ) 1 Pr(YXXX 1= | 1 , 2= ,... ) k ⎛ 1 ⎞ 1+ ⎜ ⎟ (⎝ eβ+0 βXX 1 1 + β 2 2 + ...βKK +X ⎠ ) Logit nd probita models are basically the same, the difference is in the distribution: • Logit – Cumulative standard logistic distribution (F) • Probit – Cumulative standard normal distribution (Φ) Both models provide similar results. PU/DSS/OTR It tests whether the combined effect, of all the variables in the model, is different from zero. If, for example, < 0.05 then the model have some relevant explanatory power, which does not mean it is well specified or at all correct. Logit: predicted probabilities After running the model: logit y_bin x1 x2 x3 x4 x5 x6 x7 Type predict y_bin_hat /*These are the predicted probabilities of Y=1 */ Here are the estimations for the first five cases, type: 1 x2 x3 x4 x5 x6 x7 y_bin_hatbrowse y_bin x Predicted probabilities To estimate the probability of Y=1 for the first row, replace the values of X into the logit regression equation. -
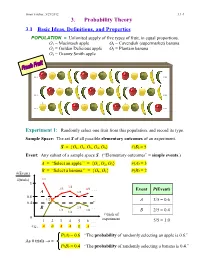
3. Probability Theory
Ismor Fischer, 5/29/2012 3.1-1 3. Probability Theory 3.1 Basic Ideas, Definitions, and Properties POPULATION = Unlimited supply of five types of fruit, in equal proportions. O1 = Macintosh apple O4 = Cavendish (supermarket) banana O2 = Golden Delicious apple O5 = Plantain banana O3 = Granny Smith apple … … … … … … Experiment 1: Randomly select one fruit from this population, and record its type. Sample Space: The set S of all possible elementary outcomes of an experiment. S = {O1, O2, O3, O4, O5} #(S) = 5 Event: Any subset of a sample space S. (“Elementary outcomes” = simple events.) A = “Select an apple.” = {O1, O2, O3} #(A) = 3 B = “Select a banana.” = {O , O } #(B) = 2 #(Event) 4 5 #(trials) 1/1 1 3/4 2/3 4/6 . Event P(Event) A 3/5 0.6 1/2 A 3/5 = 0.6 0.4 B 2/5 . 1/3 2/6 1/4 B 2/5 = 0.4 # trials of 0 experiment 1 2 3 4 5 6 . 5/5 = 1.0 e.g., A B A A B A . P(A) = 0.6 “The probability of randomly selecting an apple is 0.6.” As # trials → ∞ P(B) = 0.4 “The probability of randomly selecting a banana is 0.4.” Ismor Fischer, 5/29/2012 3.1-2 General formulation may be facilitated with the use of a Venn diagram: Experiment ⇒ Sample Space: S = {O1, O2, …, Ok} #(S) = k A Om+1 Om+2 O2 O1 O3 Om+3 O 4 . Om . Ok Event A = {O1, O2, …, Om} ⊆ S #(A) = m ≤ k Definition: The probability of event A, denoted P(A), is the long-run relative frequency with which A is expected to occur, as the experiment is repeated indefinitely. -

On Discriminative Bayesian Network Classifiers and Logistic Regression
On Discriminative Bayesian Network Classifiers and Logistic Regression Teemu Roos and Hannes Wettig Complex Systems Computation Group, Helsinki Institute for Information Technology, P.O. Box 9800, FI-02015 HUT, Finland Peter Gr¨unwald Centrum voor Wiskunde en Informatica, P.O. Box 94079, NL-1090 GB Amsterdam, The Netherlands Petri Myllym¨aki and Henry Tirri Complex Systems Computation Group, Helsinki Institute for Information Technology, P.O. Box 9800, FI-02015 HUT, Finland Abstract. Discriminative learning of the parameters in the naive Bayes model is known to be equivalent to a logistic regression problem. Here we show that the same fact holds for much more general Bayesian network models, as long as the corresponding network structure satisfies a certain graph-theoretic property. The property holds for naive Bayes but also for more complex structures such as tree-augmented naive Bayes (TAN) as well as for mixed diagnostic-discriminative structures. Our results imply that for networks satisfying our property, the condi- tional likelihood cannot have local maxima so that the global maximum can be found by simple local optimization methods. We also show that if this property does not hold, then in general the conditional likelihood can have local, non-global maxima. We illustrate our theoretical results by empirical experiments with local optimization in a conditional naive Bayes model. Furthermore, we provide a heuristic strategy for pruning the number of parameters and relevant features in such models. For many data sets, we obtain good results with heavily pruned submodels containing many fewer parameters than the original naive Bayes model. Keywords: Bayesian classifiers, Bayesian networks, discriminative learning, logistic regression 1. -

Probability Cheatsheet V2.0 Thinking Conditionally Law of Total Probability (LOTP)
Probability Cheatsheet v2.0 Thinking Conditionally Law of Total Probability (LOTP) Let B1;B2;B3; :::Bn be a partition of the sample space (i.e., they are Compiled by William Chen (http://wzchen.com) and Joe Blitzstein, Independence disjoint and their union is the entire sample space). with contributions from Sebastian Chiu, Yuan Jiang, Yuqi Hou, and Independent Events A and B are independent if knowing whether P (A) = P (AjB )P (B ) + P (AjB )P (B ) + ··· + P (AjB )P (B ) Jessy Hwang. Material based on Joe Blitzstein's (@stat110) lectures 1 1 2 2 n n A occurred gives no information about whether B occurred. More (http://stat110.net) and Blitzstein/Hwang's Introduction to P (A) = P (A \ B1) + P (A \ B2) + ··· + P (A \ Bn) formally, A and B (which have nonzero probability) are independent if Probability textbook (http://bit.ly/introprobability). Licensed and only if one of the following equivalent statements holds: For LOTP with extra conditioning, just add in another event C! under CC BY-NC-SA 4.0. Please share comments, suggestions, and errors at http://github.com/wzchen/probability_cheatsheet. P (A \ B) = P (A)P (B) P (AjC) = P (AjB1;C)P (B1jC) + ··· + P (AjBn;C)P (BnjC) P (AjB) = P (A) P (AjC) = P (A \ B1jC) + P (A \ B2jC) + ··· + P (A \ BnjC) P (BjA) = P (B) Last Updated September 4, 2015 Special case of LOTP with B and Bc as partition: Conditional Independence A and B are conditionally independent P (A) = P (AjB)P (B) + P (AjBc)P (Bc) given C if P (A \ BjC) = P (AjC)P (BjC). -

Understanding Relative Risk, Odds Ratio, and Related Terms: As Simple As It Can Get Chittaranjan Andrade, MD
Understanding Relative Risk, Odds Ratio, and Related Terms: As Simple as It Can Get Chittaranjan Andrade, MD Each month in his online Introduction column, Dr Andrade Many research papers present findings as odds ratios (ORs) and considers theoretical and relative risks (RRs) as measures of effect size for categorical outcomes. practical ideas in clinical Whereas these and related terms have been well explained in many psychopharmacology articles,1–5 this article presents a version, with examples, that is meant with a view to update the knowledge and skills to be both simple and practical. Readers may note that the explanations of medical practitioners and examples provided apply mostly to randomized controlled trials who treat patients with (RCTs), cohort studies, and case-control studies. Nevertheless, similar psychiatric conditions. principles operate when these concepts are applied in epidemiologic Department of Psychopharmacology, National Institute research. Whereas the terms may be applied slightly differently in of Mental Health and Neurosciences, Bangalore, India different explanatory texts, the general principles are the same. ([email protected]). ABSTRACT Clinical Situation Risk, and related measures of effect size (for Consider a hypothetical RCT in which 76 depressed patients were categorical outcomes) such as relative risks and randomly assigned to receive either venlafaxine (n = 40) or placebo odds ratios, are frequently presented in research (n = 36) for 8 weeks. During the trial, new-onset sexual dysfunction articles. Not all readers know how these statistics was identified in 8 patients treated with venlafaxine and in 3 patients are derived and interpreted, nor are all readers treated with placebo. These results are presented in Table 1. -

Lecture 5: Naive Bayes Classifier 1 Introduction
CSE517A Machine Learning Spring 2020 Lecture 5: Naive Bayes Classifier Instructor: Marion Neumann Scribe: Jingyu Xin Reading: fcml 5.2.1 (Bayes Classifier, Naive Bayes, Classifying Text, and Smoothing) Application Sentiment analysis aims at discovering people's opinions, emo- tions, feelings about a subject matter, product, or service from text. Take some time to answer the following warm-up questions: (1) Is this a regression or classification problem? (2) What are the features and how would you represent them? (3) What is the prediction task? (4) How well do you think a linear model will perform? 1 Introduction Thought: Can we model p(y j x) without model assumptions, e.g. Gaussian/Bernoulli distribution etc.? Idea: Estimate p(y j x) from the data directly, then use the Bayes classifier. Let y be discrete, Pn i=1 I(xi = x \ yi = y) p(y j x) = Pn (1) i=1 I(xi = x) where the numerator counts all examples with input x that have label y and the denominator counts all examples with input x. Figure 1: Illustration of estimatingp ^(y j x) From Figure ??, it is clear that, using the MLE method, we can estimatep ^(y j x) as jCj p^(y j x) = (2) jBj But there is a big problem with this method: The MLE estimate is only good if there are many training vectors with the same identical features as x. 1 2 This never happens for high-dimensional or continuous feature spaces. Solution: Bayes Rule. p(x j y) p(y) p(y j x) = (3) p(x) Let's estimate p(x j y) and p(y) instead! 2 Naive Bayes We have a discrete label space C that can either be binary f+1; −1g or multi-class f1; :::; Kg. -

Hybrid of Naive Bayes and Gaussian Naive Bayes for Classification: a Map Reduce Approach
International Journal of Innovative Technology and Exploring Engineering (IJITEE) ISSN: 2278-3075, Volume-8, Issue-6S3, April 2019 Hybrid of Naive Bayes and Gaussian Naive Bayes for Classification: A Map Reduce Approach Shikha Agarwal, Balmukumd Jha, Tisu Kumar, Manish Kumar, Prabhat Ranjan Abstract: Naive Bayes classifier is well known machine model could be used without accepting Bayesian probability learning algorithm which has shown virtues in many fields. In or using any Bayesian methods. Despite their naive design this work big data analysis platforms like Hadoop distributed and simple assumptions, Naive Bayes classifiers have computing and map reduce programming is used with Naive worked quite well in many complex situations. An analysis Bayes and Gaussian Naive Bayes for classification. Naive Bayes is manily popular for classification of discrete data sets while of the Bayesian classification problem showed that there are Gaussian is used to classify data that has continuous attributes. sound theoretical reasons behind the apparently implausible Experimental results show that Hybrid of Naive Bayes and efficacy of types of classifiers[5]. A comprehensive Gaussian Naive Bayes MapReduce model shows the better comparison with other classification algorithms in 2006 performance in terms of classification accuracy on adult data set showed that Bayes classification is outperformed by other which has many continuous attributes. approaches, such as boosted trees or random forests [6]. An Index Terms: Naive Bayes, Gaussian Naive Bayes, Map Reduce, Classification. advantage of Naive Bayes is that it only requires a small number of training data to estimate the parameters necessary I. INTRODUCTION for classification. The fundamental property of Naive Bayes is that it works on discrete value.