Exploration of Binary Protein-Protein Interactions Between Tick-Borne Flaviviruses and Ixodes Ricinus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer Et Al
USOO8440393B2 (12) United States Patent (10) Patent No.: US 8.440,393 B2 Birrer et al. (45) Date of Patent: May 14, 2013 (54) PRO-ANGIOGENIC GENES IN OVARIAN OTHER PUBLICATIONS TUMORENDOTHELIAL CELL, SOLATES Boyd (The Basic Science of Oncology, 1992, McGraw-Hill, Inc., p. (75) Inventors: Michael J. Birrer, Mt. Airy, MD (US); 379). Tomas A. Bonome, Washington, DC Tockman et al. (Cancer Res., 1992, 52:2711s-2718s).* (US); Anil Sood, Pearland, TX (US); Pritzker (Clinical Chemistry, 2002, 48: 1147-1150).* Chunhua Lu, Missouri City, TX (US) Benedict et al. (J. Exp. Medicine, 2001, 193(1) 89-99).* Jiang et al. (J. Biol. Chem., 2003, 278(7) 4763-4769).* (73) Assignees: The United States of America as Matsushita et al. (FEBS Letters, 1999, vol. 443, pp. 348-352).* Represented by the Secretary of the Singh et al. (Glycobiology, 2001, vol. 11, pp. 587-592).* Department of Health and Human Abbosh et al. (Cancer Res. Jun. 1, 2006 66:5582-55.91 and Supple Services, Washington, DC (US); The mental Figs. S1-S7).* University of MD Anderson Cancer Zhai et al. (Chinese General Practice Aug. 2008, 11(8A): 1366 Center, Houston, TX (US) 1367).* Lu et al. (Cancer Res. Feb. 15, 2007, 64(4): 1757-1768).* (*) Notice: Subject to any disclaimer, the term of this Bagnato et al., “Activation of Mitogenic Signaling by Endothelin 1 in patent is extended or adjusted under 35 Ovarian Carcinoma Cells', Cancer Research, vol. 57, pp. 1306-1311, U.S.C. 154(b) by 194 days. 1997. Bouras et al., “Stanniocalcin 2 is an Estrogen-responsive Gene (21) Appl. -

Pathogenetic Subgroups of Multiple Myeloma Patients
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector ARTICLE High-resolution genomic profiles define distinct clinico- pathogenetic subgroups of multiple myeloma patients Daniel R. Carrasco,1,2,8 Giovanni Tonon,1,8 Yongsheng Huang,3,8 Yunyu Zhang,1 Raktim Sinha,1 Bin Feng,1 James P. Stewart,3 Fenghuang Zhan,3 Deepak Khatry,1 Marina Protopopova,5 Alexei Protopopov,5 Kumar Sukhdeo,1 Ichiro Hanamura,3 Owen Stephens,3 Bart Barlogie,3 Kenneth C. Anderson,1,4 Lynda Chin,1,7 John D. Shaughnessy, Jr.,3,9 Cameron Brennan,6,9 and Ronald A. DePinho1,5,9,* 1 Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts 02115 2 Department of Pathology, Brigham and Women’s Hospital, Boston, Massachusetts 02115 3 The Donna and Donald Lambert Laboratory of Myeloma Genetics, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205 4 The Jerome Lipper Multiple Myeloma Center, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts 02115 5 Center for Applied Cancer Science, Belfer Institute for Innovative Cancer Science, Dana-Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts 6 Neurosurgery Service, Memorial Sloan-Kettering Cancer Center, Department of Neurosurgery, Weill Cornell Medical College, New York, New York 10021 7 Department of Dermatology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts, 02115 8 These authors contributed equally to this work. 9 Cocorresponding authors: [email protected] (C.B.); [email protected] (J.D.S.); [email protected] (R.A.D.) *Correspondence: [email protected] Summary To identify genetic events underlying the genesis and progression of multiple myeloma (MM), we conducted a high-resolu- tion analysis of recurrent copy number alterations (CNAs) and expression profiles in a collection of MM cell lines and out- come-annotated clinical specimens. -

Susceptibility to Glaucoma: Differential Comparison of the Astrocyte
Open Access Research2008LukasetVolume al. 9, Issue 7, Article R111 Susceptibility to glaucoma: differential comparison of the astrocyte transcriptome from glaucomatous African American and Caucasian American donors Thomas J Lukas¤*, Haixi Miao¤†, Lin Chen†, Sean M Riordan†, Wenjun Li†, Andrea M Crabb†, Alexandria Wise‡, Pan Du§, Simon M Lin§ and M Rosario Hernandez† Addresses: *Department of Molecular Pharmacology and Biological Chemistry, Feinberg School of Medicine, Northwestern University, E Chicago Ave, Chicago, IL 60611 USA. †Department of Ophthalmology, Feinberg School of Medicine, Northwestern University, E Chicago Ave, Chicago, IL 60611 USA. ‡Department of Biology, City College of New York, Convent Ave, New York, NY 10031, USA. §Robert H Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, E Chicago Ave, Chicago, IL 60611 USA. ¤ These authors contributed equally to this work. Correspondence: Thomas J Lukas. Email: [email protected] Published: 9 July 2008 Received: 9 May 2008 Revised: 18 June 2008 Genome Biology 2008, 9:R111 (doi:10.1186/gb-2008-9-7-r111) Accepted: 9 July 2008 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2008/9/7/R111 © 2008 Lukas et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Epidemiological and genetic studies indicate that ethnic/genetic background plays an important role in susceptibility to primary open angle glaucoma (POAG). -

Identification of Dysregulated Genes in Rheumatoid Arthritis Based on Bioinformatics Analysis
Identification of dysregulated genes in rheumatoid arthritis based on bioinformatics analysis Ruihu Hao1,*, Haiwei Du2,*, Lin Guo1, Fengde Tian1, Ning An1, Tiejun Yang3, Changcheng Wang1, Bo Wang1 and Zihao Zhou1 1 Department of Orthopedics, Affiliated Zhongshan Hospital of Dalian University, Dalian, China 2 Department of Bioinformatics, Beijing Medintell Biomed Co., Ltd, Beijing, China 3 Department of Orthopedics, Affiliated Hospital of BeiHua University, Jilin, China * These authors contributed equally to this work. ABSTRACT Background. Rheumatoid arthritis (RA) is a chronic auto-inflammatory disorder of joints. The present study aimed to identify the key genes in RA for better understanding the underlying mechanisms of RA. Methods. The integrated analysis of expression profiling was conducted to identify differentially expressed genes (DEGs) in RA. Moreover, functional annotation, protein– protein interaction (PPI) network and transcription factor (TF) regulatory network construction were applied for exploring the potential biological roles of DEGs in RA. In addition, the expression level of identified candidate DEGs was preliminarily detected in peripheral blood cells of RA patients in the GSE17755 dataset. Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to validate the expression levels of identified DEGs in RA. Results. A total of 378 DEGs, including 202 up- and 176 down-regulated genes, were identified in synovial tissues of RA patients compared with healthy controls. DEGs were significantly enriched in -

Discovery, Expression Profiling, and Evolutionary Analysis Of
DISCOVERY, EXPRESSION PROFILING, AND EVOLUTIONARY ANALYSIS OF CYNODON EXPRESSED SEQUENCE TAGS by CHANGSOO KIM (Under the Direction of Andrew H. Paterson) ABSTRACT Bermudagrass (Cynodon dactylon) is a major turfgrass species for sports fields, lawns, parks, golf courses, and general utility turfs in tropical and subtropical regions. Despite its ecological importance, much of its study has been dependent upon classical approaches. Information about Bermudagrass at the molecular level has been deficient although molecular information for other plants has been accumulated for the last two dacades. In the current study, we constructed a normalized cDNA library from leaf tissue of Bermudagrass in order to expand our knowledge of its transcriptome. We sequenced and annotated 15,588 expressed sequence tags (ESTs), which were deposited in the National Center for Biotechnology Information (NCBI) to be shared with other scientists. We also conducted cDNA array hybridization (macroarray) to profile genes responding to drought stress. A total of 120 and 69 genes were identified as up- and down-regulated, respectively. BLASTX annotation suggested that up-regulated genes may be involved in osmotic adjustment, signal transduction pathways, protein repair systems, and removal of toxins, while down-regulated genes were mostly related to basic plant metabolism such as photosynthesis and glycolysis. Using the cDNA sequences, we performed a comparative genomic study to gain new insight into the evolution of Bermudagrass. Results suggested that the common ancestor of the grass family experienced a whole genome duplication event at ca. 50.0 ~ 65.4 million years ago (MYA), before the divergence of the PACC and BEP clades at ca. 42.3 ~ 50.0 MYA. -

Platform Abstracts
American Society of Human Genetics 65th Annual Meeting October 6–10, 2015 Baltimore, MD PLATFORM ABSTRACTS Wednesday, October 7, 9:50-10:30am Abstract #’s Friday, October 9, 2:15-4:15 pm: Concurrent Platform Session D: 4. Featured Plenary Abstract Session I Hall F #1-#2 46. Hen’s Teeth? Rare Variants and Common Disease Ballroom I #195-#202 Wednesday, October 7, 2:30-4:30pm Concurrent Platform Session A: 47. The Zen of Gene and Variant 15. Update on Breast and Prostate Assessment Ballroom III #203-#210 Cancer Genetics Ballroom I #3-#10 48. New Genes and Mechanisms in 16. Switching on to Regulatory Variation Ballroom III #11-#18 Developmental Disorders and 17. Shedding Light into the Dark: From Intellectual Disabilities Room 307 #211-#218 Lung Disease to Autoimmune Disease Room 307 #19-#26 49. Statistical Genetics: Networks, 18. Addressing the Difficult Regions of Pathways, and Expression Room 309 #219-#226 the Genome Room 309 #27-#34 50. Going Platinum: Building a Better 19. Statistical Genetics: Complex Genome Room 316 #227-#234 Phenotypes, Complex Solutions Room 316 #35-#42 51. Cancer Genetic Mechanisms Room 318/321 #235-#242 20. Think Globally, Act Locally: Copy 52. Target Practice: Therapy for Genetic Hilton Hotel Number Variation Room 318/321 #43-#50 Diseases Ballroom 1 #243-#250 21. Recent Advances in the Genetic Basis 53. The Real World: Translating Hilton Hotel of Neuromuscular and Other Hilton Hotel Sequencing into the Clinic Ballroom 4 #251-#258 Neurodegenerative Phenotypes Ballroom 1 #51-#58 22. Neuropsychiatric Diseases of Hilton Hotel Friday, October 9, 4:30-6:30pm Concurrent Platform Session E: Childhood Ballroom 4 #59-#66 54. -

Characterizing Embryonic Gene Expression Patterns in the Mouse
Downloaded from genome.cshlp.org on September 30, 2015 - Published by Cold Spring Harbor Laboratory Press Letter Characterizing Embryonic Gene Expression Patterns in the Mouse Using Nonredundant Sequence-Based Selection Rita Sousa-Nunes,1,10 Amer Ahmed Rana,1,10,7 Ross Kettleborough,1,10 Joshua M. Brickman,1,8 Melanie Clements,1 Alistair Forrest,2 Sean Grimmond,2 Philip Avner,3 James C. Smith,4,11 Sally L. Dunwoodie,1,5,6,11 and Rosa S.P. Beddington1,9 1Division of Mammalian Development, National Institute for Medical Research, The Ridgeway, London NW7 1AA, United Kingdom; 2Institute of Molecular Bioscience, University of Queensland, 4072 Australia; 3Unite´Ge´ne´tique Mole´culaire Murine, Institut Pasteur, 75015 Paris, France; 4Wellcome Trust/Cancer Research UK Institute and Department of Zoology, University of Cambridge, Cambridge CB2 1QR, United Kingdom; 5Developmental Biology Program, Victor Chang Cardiac Research Institute, Darlinghurst, 2010, Australia; 6Department of Biotechnology and Biomolecular Sciences, University of New South Wales, Kensington, NSW 2033, Australia This article investigates the expression patterns of 160 genes that are expressed during early mouse development. The cDNAs were isolated from 7.5 d postcoitum (dpc) endoderm, a region that comprises visceral endoderm (VE), definitive endoderm, and the node–tissues that are required for the initial steps of axial specification and tissue patterning in the mouse. To avoid examining the same gene more than once, and to exclude potentially ubiquitously expressed housekeeping genes, cDNA sequence was derived from 1978 clones of the Endoderm library. These yielded 1440 distinct cDNAs, of which 123 proved to be novel in the mouse. -
Wo 2010/040571 A2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date 15 April 2010 (15.04.2010) WO 2010/040571 A2 (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C12N 15/11 (2006.01) kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, (21) International Application Number: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, PCT/EP2009/00743 1 DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (22) International Filing Date: HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, 12 October 2009 (12.10.2009) KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, (25) Filing Language: English NO, NZ, OM, PE, PG, PH, PL, PT, RO, RS, RU, SC, SD, (26) Publication Language: English SE, SG, SK, SL, SM, ST, SV, SY, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 08075816.2 10 October 2008 (10.10.2008) DE (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicant (for all designated States except US): GM, KE, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, ZM, FRAUNHOFER-GESELLSCHAFT ZUR ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, FORDERUNG DER ANGEWANDTEN TM), European (AT, BE, BG, CH, CY, CZ, DE, DK, EE, FORSCHUNG E.V. -
Exploration of Binary Protein–Protein Interactions Between Tick-Borne Flaviviruses and Ixodes Ricinus
Lemasson et al. Parasites Vectors (2021) 14:144 https://doi.org/10.1186/s13071-021-04651-3 Parasites & Vectors RESEARCH Open Access Exploration of binary protein–protein interactions between tick-borne faviviruses and Ixodes ricinus Manon Lemasson1, Grégory Caignard1, Yves Unterfnger1, Houssam Attoui1, Lesley Bell‑Sakyi2, Edouard Hirchaud3, Sara Moutailler4, Nicholas Johnson5, Damien Vitour1, Jennifer Richardson1 and Sandrine A. Lacour1* Abstract Background: Louping ill virus (LIV) and tick‑borne encephalitis virus (TBEV) are tick‑borne faviviruses that are both transmitted by the major European tick, Ixodes ricinus. Despite the importance of I. ricinus as an arthropod vector, its capacity to acquire and subsequently transmit viruses, known as vector competence, is poorly understood. At the molecular scale, vector competence is governed in part by binary interactions established between viral and cellular proteins within infected tick cells. Methods: To investigate virus‑vector protein–protein interactions (PPIs), the entire set of open reading frames for LIV and TBEV was screened against an I. ricinus cDNA library established from three embryonic tick cell lines using yeast two‑hybrid methodology (Y2H). PPIs revealed for each viral bait were retested in yeast by applying a gap repair (GR) strategy, and notably against the cognate protein of both viruses, to determine whether the PPIs were specifc for a single virus or common to both. The interacting tick proteins were identifed by automatic BLASTX, and in silico analy‑ ses were performed to expose the biological processes targeted by LIV and TBEV. Results: For each virus, we identifed 24 diferent PPIs involving six viral proteins and 22 unique tick proteins, with all PPIs being common to both viruses. -

COPI Activity Coupled with Fatty Acid Biosynthesis Is Required for Viral Replication
COPI Activity Coupled with Fatty Acid Biosynthesis Is Required for Viral Replication Sara Cherry1*, Amit Kunte2, Hui Wang3, Carolyn Coyne4, Robert B. Rawson2, Norbert Perrimon3 1 University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, United States of America, 2 University of Texas Southwestern Medical Center, Dallas, Texas, United States of America, 3 Harvard Medical School, Howard Hughes Medical Institute, Boston, Massachusetts, United States of America, 4 Children’s Hospital of Pennsylvania, Philadelphia, Pennsylvania, United States of America During infection by diverse viral families, RNA replication occurs on the surface of virally induced cytoplasmic membranes of cellular origin. How this process is regulated, and which cellular factors are required, has been unclear. Moreover, the host–pathogen interactions that facilitate the formation of this new compartment might represent critical determinants of viral pathogenesis, and their elucidation may lead to novel insights into the coordination of vesicular trafficking events during infection. Here we show that in Drosophila cells, Drosophila C virus remodels the Golgi apparatus and forms a novel vesicular compartment, on the surface of which viral RNA replication takes place. Using genome-wide RNA interference screening, we found that this step in the viral lifecycle requires at least two host encoded pathways: the coat protein complex I (COPI) coatamer and fatty acid biosynthesis. Our results integrate, clarify, and extend numerous observations concerning the cell biology of viral replication, allowing us to conclude that the coupling of new cellular membrane formation with the budding of these vesicles from the Golgi apparatus allows for the regulated generation of this new virogenic organelle, which is essential for viral replication. -
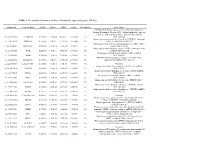
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137. -

Data Set 1. Biological Analysis of the Genes Found to Be Significant in the Endotoxin Study
Data Set 1. Biological analysis of the genes found to be significant in the endotoxin study. Pages 2 – 105: Q values, gene names and annotation on the genes significant at FDR = 0.1%. Pages 106 – 127: Global functional analysis of the down-regulated genes. Pages 128 – 169: Global functional analysis of the up-regulated genes. Probe Set q-value Direction Gene Annotation 117_at 9.60E-05 up HSPA6 heat shock 70kDa protein 6 (HSP70B') 1405_i_at 2.00E-06 down CCL5 chemokine (C-C motif) ligand 5 PRP8 pre-mRNA processing factor 8 200000_s_at 2.50E-05 down PRPF8 homolog (yeast) PRP8 pre-mRNA processing factor 8 200000_s_at 0.000712 down PRPF8 homolog (yeast) 200001_at 0.000658 down CAPNS1 calpain, small subunit 1 200002_at 2.00E-06 down RPL35 ribosomal protein L35 200002_at 2.00E-06 down RPL35 ribosomal protein L35 200003_s_at 1.10E-05 down RPL28 ribosomal protein L28 200003_s_at 1.10E-05 down RPL28 ribosomal protein L28 eukaryotic translation initiation factor 3, 200005_at 2.00E-06 down EIF3S7 subunit 7 zeta, 66/67kDa eukaryotic translation initiation factor 3, 200005_at 2.00E-06 down EIF3S7 subunit 7 zeta, 66/67kDa Parkinson disease (autosomal 200006_at 4.70E-05 down PARK7 recessive, early onset) 7 Parkinson disease (autosomal 200006_at 8.70E-05 down PARK7 recessive, early onset) 7 200008_s_at 5.40E-05 up GDI2 GDP dissociation inhibitor 2 200008_s_at 0.000361 up GDI2 GDP dissociation inhibitor 2 200009_at 0.000171 down GDI2 GDP dissociation inhibitor 2 200010_at 2.00E-06 down RPL11 ribosomal protein L11 200010_at 2.00E-06 down RPL11 ribosomal