VISUAL Network Analysis Reveals the Role of BEH3 As a Stabilizer in The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
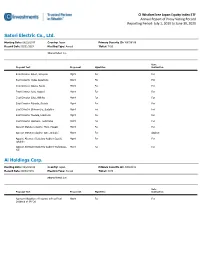
Proxy Voting Record Reporting Period: July 1, 2019 to June 30, 2020
CI WisdomTree Japan Equity Index ETF Annual Report of Proxy Voting Record Reporting Period: July 1, 2019 to June 30, 2020 Satori Electric Co., Ltd. Meeting Date: 08/22/2019 Country: Japan Primary Security ID: J69736106 Record Date: 05/31/2019 Meeting Type: Annual Ticker: 7420 Shares Voted: 800 Vote Proposal Text Proponent Mgmt Rec Instruction Elect Director Satori, Hiroyuki Mgmt For For Elect Director Ueda, Kazutoshi Mgmt For For Elect Director Obara, Naoki Mgmt For For Elect Director Aoki, Yasushi Mgmt For For Elect Director Sato, Akihiko Mgmt For For Elect Director Fukuda, Shuichi Mgmt For For Elect Director Shimomura, Sadahiro Mgmt For For Elect Director Tawada, Hidetoshi Mgmt For For Elect Director Iwanami, Toshimitsu Mgmt For For Appoint Statutory Auditor Mogi, Masaki Mgmt For For Appoint Statutory Auditor Sato, Shinichi Mgmt For Against Appoint Alternate Statutory Auditor Suzuki, Mgmt For For Takahiro Appoint Alternate Statutory Auditor Yoshimasu, Mgmt For For Yuji Ai Holdings Corp. Meeting Date: 09/26/2019 Country: Japan Primary Security ID: J0060P101 Record Date: 06/30/2019 Meeting Type: Annual Ticker: 3076 Shares Voted: 100 Vote Proposal Text Proponent Mgmt Rec Instruction Approve Allocation of Income, with a Final Mgmt For For Dividend of JPY 20 CI WisdomTree Japan Equity Index ETF Proxy Voting Record | July 1, 2019 to June 30, 2020 Nippon Koei Co., Ltd. Meeting Date: 09/26/2019 Country: Japan Primary Security ID: J34770107 Record Date: 06/30/2019 Meeting Type: Annual Ticker: 1954 Shares Voted: 100 Vote Proposal Text Proponent -

Univerzita Karlova Pedagogická Fakulta BAKALÁŘSKÁ PRÁCE 2018
Univerzita Karlova Pedagogická fakulta BAKALÁŘSKÁ PRÁCE 2018 Pihávková Natálie Univerzita Karlova Pedagogická fakulta Katedra tělesné výchovy BAKALÁŘSKÁ PRÁCE Vznik a rozvoj bojového sportu MMA v České republice a v USA Establishment and development of MMA combat sport in the Czech republic and the USA Natálie Pihávková Vedoucí práce: PhDr. Martin Dlouhý, PhD. Studijní program: B7507 Specializace v pedagogice Studijní obor: Tělesná výchova a sport se zaměřením na vzdělávání - Biologie, geologie a environmentalistika se zaměřením na vzdělávání 2018 Prohlášení Prohlašuji, že jsem bakalářskou práci na téma Vznik a rozvoj bojového sportu MMA v České republice a v USA vypracovala pod vedením vedoucího práce samostatně za použití v práci uvedených pramenů a literatury. Dále prohlašuji, že tato práce nebyla využita k získání jiného nebo stejného titulu. V Praze dne 19.4.2018 Poděkování Tímto bych ráda poděkovala všem, kteří mi byli vždy oporou během vytváření mé bakalářské práce. Zvláštní poděkování patří zejména vedoucímu práce panu PhDr. Martinu Dlouhému, PhD., který mi poskytl cenné rady a nápady. Dále bych ráda poděkovala Jakubu Šnebergerovi a Zdeňku Vítovi za spolupráci při vytváření fotek obsažených v práci. A v neposlední řadě i Viktorovi Peštovi za spolupráci během rozhovoru. ANOTACE Základem bakalářské práce je popsat vznik a rozvoj MMA jak v České republice, tak v USA. V práci je věnována pozornost zejména historii MMA, ale najdeme zde i organizace věnující se tomuto sportu a nejlepší zápasníky z obou zemí. Teoretická část je rozdělena na tři velké kapitoly, ve kterých je MMA obecně, MMA v České republice a MMA v USA. Součástí bakalářské práce je i řízený rozhovor se zápasníkem, který si prošel nejen zápasením v České republice, ale i ve Spojených státech. -

Mixed Martial Arts 1 Mixed Martial Arts
Mixed martial arts 1 Mixed martial arts Mixed Martial Arts Patrick Barry (Blue shorts) and Mirko Filipović (Checkered shorts) in the co-main event of UFC 115 in Vancouver, British Columbia, Canada. Also known as Vale Tudo, No Holds Barred (NHB), Cage Fighting, Ultimate Fighting, Pride Fighting, Sougo Kakutogi Focus Various Hardness Full contact Olympic sport No Mixed martial arts (MMA), popularly known as cage fighting or ultimate fighting is a full contact combat sport that allows a wide variety of fighting techniques and skills, from a mixture of other combat sports, to be used in competitions. The rules allow the use of both striking as well as grappling techniques, both while standing and while on the ground. Such competitions allow fighters of different backgrounds to compete. The roots of modern mixed martial arts can be traced back to various mixed style contests that took place throughout Europe, Japan and the Pacific Rim during the early 1900s. The combat sport of Vale Tudo that had developed in Brazil from the 1920s was brought to the United States by the Gracie family in 1993 with the founding of the Ultimate Fighting Championship. Professional MMA events had also been held in Japan by Shooto starting back in 1989. In due course the more dangerous Vale Tudo style bouts of the early UFCs were made safer with the implementation of additional rules, leading to the popular regulated form of MMA seen today. Originally promoted as a competition with the intention of finding the most effective martial arts for real unarmed combat situations, -

Conference Agenda
PROGRAM SUNDAY, November 3—6:00 PM Welcome Reception SUNDAY, November 3—7:00 PM PLENARY TALK Evolution of chloroplasts and mitochondria—A new look through a window of the cellular system, protein targeting mechanism Inhwan Hwang, Doong Wook Lee, Junho Lee [35’+10’] Presenter affiliation: Pohang University of Science and Technology, Pohang, South Korea. 1 MONDAY, November 4—9:00 AM SESSION 1 EMBRYOGENESIS Chairperson: Dolf Weijers, Wageningen University, Wageningen, the Netherlands A conserved biochemical paradigm underlies cell polarity across multicellular kingdoms Maritza van Dop, Marc Fiedler, Sumanth Mutte, Jeroen de Keijzer, Lisa Olijslager, Marcel Janson, Mariann Bienz, Dolf Weijers [20’+10’] Presenter affiliation: Wageningen University, Wageningen, the Netherlands. 2 Small RNA functions in Arabidopsis embryos Michael D. Nodine [20’+10’] Presenter affiliation: Gregor Mendel Institute, Vienna, Austria. 3 Modulation of cellular pluripotency by cell proliferation Yoo-Sun Noh [20’+10’] Presenter affiliation: Seoul National University, Seoul, South Korea. 4 Coffee Break v Initiation of the shoot meristem stem cells in Arabidopsis thaliana Wen Gong, Thomas Laux [20’+10’] Presenter affiliation: Signalling Research Centres BIOSS and CIBSS, University of Freiburg, Germany; Sino-German Joint Research Center on Agricultural Biology, Taian, China. 5 An excision-triggered ultradian rhythm positively regulates de novo root regeneration via ABA signaling in Arabidopsis leaves Vu Thi Quy, Kitae Song, Hyo Jung Kim, Sungjin Park, Gayoung Seo, Hong Gil Nam, Sunghyun Hong [10’+5’] Presenter affiliation: IBS (Institute for Basic Science), Daegu, South Korea. 6 De novo root regeneration—From wounding to stem cell fate transition Lin Xu [10’+5’] Presenter affiliation: Chinese Academy of Sciences, Institute of Plant Physiology and Ecology, Shanghai, China. -

Plant Peptide Helps Roots to Branch out in the Right Places 21 January 2019
Plant peptide helps roots to branch out in the right places 21 January 2019 Koichi Fujimoto (Osaka University) and Assistant Professor Yuki Kondo (the University of Tokyo). Plant root systems are mainly shaped by the lateral roots that grow from tissue inside the existing roots. These roots form from "lateral root founder cells" that are positioned at regularly-spaced intervals at a distance from the meristem tissue (tissue responsible for growth). Previous studies using Arabidopsis plants showed that lateral root founder cells are made from sites where there is high response to the chemical auxin, and indicated that transcription factor LBD16 induced by auxin may inhibit the cells near lateral root founder cells from forming roots. A: Arabidopsis wild-type (left) and TOLS2 overexpression type (right). 10-day growth. Scale is 1 cm.B: The expression of DR5:LUC gene in the roots of Arabidopsis wild-type (left) and TOLS2 overexpressor (right). White arrows indicate lateral root founder cells. The scale is 1 cm. Credit: Kobe University How do plants space out their roots? A Japanese research team has identified a peptide and its receptor that help lateral roots to grow with the right spacing. The findings were published on December 20, 2018 in the online edition of Developmental Cell. The team was led by Professor Hidehiro Fukaki (Graduate School of Science, Kobe University), Researcher Koichi Toyokura (currently JSPS Research Fellow at Osaka University) and Project Assistant Professor Tatsuaki Goh (currently Assistant Professor at the Nara Institute of Science and Technology) in collaboration with Professor Yoshikatsu Matsubayashi and Assistant Professor Hidefumi Shinohara (both from Nagoya University) and other researchers from the Nara Institute of Science and Technology, Associated Professor 1 / 3 number of lateral root founder cells and lateral roots decreased (figure 2). -

Best Engine Vol.8
Best Engine Vol. 8 Special Feature Understanding and Leveraging the Transformation Taking Place in Silicon Valley Kenji Kushida Research Scholar, Shorenstein Asia-Pacific Research Center, Stanford University Project Leader, Stanford Silicon Valley ‒ New Japan Project Best Engine Vol. 8 CONTENTS Cover photo by Masataka Nakano The Four Seasons of IT 3 The Agile Tokyo, a dedicated Transformation and T-shirts space for agile development opened in Satoshi Kikuchi Tokyo. This space opens President & CEO up connections with customers and partners for fast, optimized application development through 4 Special Feature communication. The Agile Tokyo also collaborates Understanding and Leveraging with agile development spaces at the Toyota and the Transformation Taking Place Nagoya Offices. in Silicon Valley Special Interview Notice Research Scholar, Shorenstein Asia-Pacific Research Center, Stanford University Kenji Kushida Project Leader, Stanford Silicon Valley ‒ New Japan Project Upcoming Masataka Nakano 12 CTC Spreads Its Business Worldwide Photo Exhibit“ Tokyo” Extending the Global Reach of the CTC Group Tokyo Photographic Art Museum November 23 (Sat/Holiday), 2019 - January 26 (Sun), 2020 IT Terminology 16 Photographer Masataka esports Nakano whose work appears on the cover photos of Best Silicon Valley Report Engine has been 18 photographing the Business Transformation in the Digital Era international city of Tokyo from his own unique Teppei Tsuchikawa perspective for over three Deputy General Manager, Silicon Valley Office, ICT & Financial Business Division decades. Featuring Tokyo ITOCHU International Inc. Nobody, Tokyo Windows, and Tokyo Float, a culmination of Golf Digest Editorial̶Practical Golf Theory for Mental Toughness 20 his photography including new “Three Ways” to Tackle Your Weak Hole and unpublished works will be on exhibit. -

Spring 2013 Fall 2014 Volume 18, No. 1
FallSpring 2014 2013 Volume 18, No. 1 Newsletter of the Center for Japanese Studies School of Pacific and Asian Studies University of Hawai‘i at Mānoa Please submit materials to [email protected] Visit our website at www.hawaii.edu/cjs Director’s Message: Good News Regent Randy Moore Tours SPAS “Four More Years” for Our National Resource Center for East Asia & Foreign Language and Area Studies Fellowships, 2014-2018 Thanks to the excellence of our faculties in Asian and Pa- cific languages and area studies, SPAS again won funding from the Title VI International and Foreign Language Education program of the U.S. Department of Education. This means our National Resource Center for East Asia can continue to build model curriculum in Chinese, Korean, and Japanese lan- SPAS students and faculty welcomed UH Board of guages and area studies. Our next NRCEA projects will accel- Regents Chairman Randolph G. Moore on December 4, erate student skills in Japanese and Korean classes, add two 2014. Regent Moore is in the back row, 4th from the semesters of fifth year Chinese, design a course on Asia-Pacific right, with SPAS Dean R.A. Sutton. Globalization, and work with our College of Education in fu- UHM Students Travel to Japan ture teacher training. The NRCEA will help Hamilton Library add materials in Japanese, Chinese, and Korean. Most impor- The Japan Foundation’s “KAKEHASHI Project -The tantly, FLAS fellowships will continue to support students Bridge for Tomorrow” sponsored 23 UHM students and 2 studying Chinese, Korean, and Japanese languages. Under- UHM faculty leaders, Lonny Carlile and Gladys Nakahara, graduate and graduate students can apply on STAR using the on a ten-day cultural study tour to Japan, June 23-July 3, Keyword “FLAS.” A new provision in the FLAS regulations 2014. -

MMA Encyclopedia / Jonathan Snowden and Kendall Shields
00MMAEncycl_i-iv__ 02/09/10 3:54 PM Page i ECW Press 00MMAEncycl_i-iv__ 02/09/10 3:54 PM Page ii Copyright © Jonathan Snowden and Kendall Shields, 2010 Published by ECW Press 2120 Queen Street East, Suite 200, Toronto, Ontario, Canada m4e 1e2 416-694-3348 [email protected] All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form by any process — electronic, mechanical, photocopying, recording, or other- wise — without the prior written permission of the copyright owners and ECW Press. The scanning, uploading, and distribution of this book via the Internet or via any other means without the permission of the publisher is illegal and punishable by law. Please purchase only authorized electronic editions, and do not participate in or encourage electronic piracy of copyrighted materials. Your support of the authors’ rights is appreciated. library and archives canada cataloguing in publication Snowden, Jonathan, 1975- The MMA encyclopedia / Jonathan Snowden and Kendall Shields. Includes bibliographical references. isbn 978-1-55022-923-3 1. Mixed martial arts--Encyclopedias. i. Shields, Kendall ii. Title. gv1102.7.m59s65 2010 796.81503 c2010-901256-9 Developing Editor: Michael Holmes Cover Design: Dave Gee Text Design: Tania Craan Color Section Design: Rachel Ironstone Typesetting: Gail Nina Photos copyright © Peter Lockley, 2010 Printing: Solisco Tri-Graphic 1 2 3 4 5 The publication of The MMA Encyclopedia has been generously supported by the Government of Ontario through Ontario Book Publishing Tax Credit, by the OMDC Book Fund, an initiative of the Ontario Media Development Corporation, and by the Government of Canada through the Canada Book Fund. -

All Weights of UFC Champions, Men and Women, & the Ultimate Fighter & Contender Series
All Weights of UFC Champions, Men and Women, & The Ultimate Fighter & Contender Series Men UFC Champions Men's Heavyweight Champion Loser Date Defenses Event Heavyweight 206‐265 Stipe Miočić Daniel Cormier August 17, 2019 0 UFC 241 Heavyweight 206‐265 Daniel Cormier Derrick Lewis November 3, 2018 1 UFC 230 Heavyweight 206‐265 Daniel Cormier Stipe Miočić July 7, 2018 0 UFC 226 Heavyweight 206‐265 Stipe Miočić Francis Ngannou January 20, 2018 3 UFC 220 Heavyweight 206‐265 Stipe Miočić Junior dos Santos May 13, 2017 2 UFC 211 Heavyweight 206‐265 Stipe Miočić Alistair Overeem September 10, 2016 1 UFC 203 Heavyweight 206‐265 Stipe Miočić Fabrício Werdum May 14, 2016 0 UFC 198 Heavyweight 206‐265 Fabrício Werdum Cain Velasquez June 13, 2015 0 UFC 188 Heavyweight 206‐265 Fabrício Werdum (IC) Mark Hunt November 15, 2014 0 UFC 180 (IC) Interim UFC Heavyweight Champion due to injury to Cain Velasquez Heavyweight 206‐265 Cain Velasquez Junior dos Santos October 19, 2013 2 UFC 166 Heavyweight 206‐265 Cain Velasquez Antonio Silva May 25, 2013 1 UFC 160 Heavyweight 206‐265 Cain Velasquez Junior dos Santos December 29, 2012 0 UFC 155 Heavyweight 206‐265 Junior dos Santos Frank Mir May 26, 2012 1 UFC 146 Heavyweight 206‐265 Junior dos Santos Cain Velasquez November 12, 2011 0 UFC Fox 1 Heavyweight 206‐265 Cain Velasquez Brock Lesnar October 23, 2010 0 UFC 121 Heavyweight 206‐265 Brock Lesnar Shane Carwin (IC) July 3, 2010 0 UFC 116 Heavyweight 206‐265 Shane Carwin (IC) Frank Mir March 27, 2010 0 UFC 111 Heavyweight 206‐265 Brock Lesnar (IC) Frank Mir -

« Arts Martiaux Mixtes » Couramment Appelés MMA
Mission parlementaire sur la pratique des « Arts Martiaux Mixtes » couramment appelés MMA Monsieur Patrick VIGNAL, Député Monsieur Jacques GROSPERRIN, Sénateur Parlementaires en mission auprès de Monsieur Patrick Kanner, ministre de la Ville, de la Jeunesse et des Sports, et Monsieur Thierry Braillard, Secrétaire d’État chargé des Sports, Lettre signée le 7 avril 2016 par Monsieur Manuel Valls, Premier ministre. Mission parlementaire sur la pratique des « Arts Martiaux Mixtes » SOMMAIRE INTRODUCTION ...................................................................................................................................4 MÉTHODOLOGIE ET CADRAGE DE LA MISSION ............................................................................ 6 1. La pratique des Arts Martiaux Mixtes en France .........................................................................7 a. Les origines du Mixed Martial Arts (MMA) 7 i. La définition et l’histoire du MMA 7 ii. Le MMA sur le plan international 7 iii. La recommandation R.99-11 du Conseil de l’Europe du 22 avril 1999 9 iv. La position du conseil supérieur de l’audiovisuel (CSA) 10 v. Les différentes législations sur le MMA 10 vi. L’article L331-2 du code du sport 12 vii. La position du ministère chargé des Sports 13 b. Les particularités de l’arrivée du MMA en France 16 i. Le développement des nouvelles pratiques des arts martiaux mixtes et des sports de combat 16 ii. Les pratiques des arts martiaux mixtes dites amateurs en France 17 iii. Les pratiques du MMA dites professionnelles 18 CONCLUSION DE LA PREMIÈRE PARTIE ....................................................................................... 21 2. Diagnostic et évaluation des enjeux de la prise en compte d’une pratique sportive émergente des arts martiaux et des sports de combat existants �������������������������������������������� 22 a. Les effets de la société sur les pratiques sportives des arts martiaux et des sports de combat 22 b. -

Wednesday, September 11 (AM)
Wednesday, September 11 (AM) Room A Room B Room C 9:00 Special Lecture I : The Japanese Photochemistry Association Lectureship Award (2013) KOSHIMA, Hideko (Ehime Univ.) “Solid-state Organic Photochemistry: from Motion of Molecules to Mechanical Motion of Crystals” 9:35 [Room A] 9:40 1A01 1B01 1C01 Conformational Orientation of Individual Analysis of Photophysical Properties of Single Polymer Chain Studied Mechano-Response of a Cultured T-Shaped π-Conjugated by Super-Resolution Optical Myoblast by Femtosecond Molecules Based on Idazole Group Microscopy (Kyoto Univ.) AOKI, Laser-Induced Impulsive Force (NAIST) INOUCHI, Toshifumi; Hiroyuki; KURODA, Taiki (NAIST) HOSOKAWA, Yoichiroh; NAKASHIMA, Takuya; KAWAI, SAKAGUCHI, Sayaka; IINO, Tsuyoshi Takanori 10:00 1A02 1B02 1C02 Fabrication of Fluorescently Template that Grows to Protein Solid-State cistrans Labeled Diatom Silica Shells Crystal Produced by Photoisomerization of and Optical Data Storage Photochemical Reaction (Gunma Azobenzenes (Univ. of Tokyo, (Yamagata Univ., Univ.) OKUTSU, Tetsuo; Hokkaido Univ.) UCHIDA, Ayano; JST-PRESTO) HOTTA, KUROIWA, Takashi; TAKASE, TOYOTA, Taro; OGAWA, Jun-ichi; HORIUCHI, Yuki Yuta; IIZUKA, Shiori; UTSUMI, Keiichiro; HARADA, Jun Maiko; HORIUCHI, Hiroaki 10:20 1A03 1B03 1C03 In Situ Sensing of Intracellular Intracellular Oxygen Photochromism of pH Using Autofluorescence Measurements Based on the [2.2]Paracyclophane-Bridged Lifetime Imaging Phosphorescence Lifetime of Bis(imidazole dimer) (Aoyama (Hokkaido Univ.) Iridium Complexes (Gunma Univ.) Gakuin Univ., -

Generation of Enterocyte-Like Cells with Pharmacokinetic Functions from Human
DMD Fast Forward. Published on February 3, 2015 as DOI: 10.1124/dmd.114.062604 This article has not been copyedited and formatted. The final version may differ from this version. DMD #62604 Title Generation of Enterocyte-Like Cells with Pharmacokinetic Functions from Human Induced Pluripotent Stem Cells Using Small-Molecule Compounds Authors Takahiro Iwao, Nao Kodama, Yuki Kondo, Tomoki Kabeya, Katsunori Nakamura, Takashi Horikawa, Takuro Niwa, Kouichi Kurose, and Tamihide Matsunaga Downloaded from Department of Clinical Pharmacy, Graduate School of Pharmaceutical Sciences, Nagoya City University, Nagoya 467-8603, Japan (T.I., N.K., Y.K., K.N., T.M.) dmd.aspetjournals.org Educational Research Center for Clinical Pharmacy, Faculty of Pharmaceutical Sciences, Nagoya City University, Nagoya 467-8603, Japan (T.I., T.K., K.N., T.M.) DMPK Research Laboratory, Mitsubishi Tanabe Pharma Corporation, Toda, Saitama at ASPET Journals on October 1, 2021 335-8505, Japan (T.H., T.N.) Research & Development Department, Japan Bioindustry Association, Tokyo 104-0032, Japan (T.N.) The Graduate School of Marine Science and Technology, Tokyo University of Marine Science and Technology, Tokyo 108-8477, Japan (K.K.) 1 DMD Fast Forward. Published on February 3, 2015 as DOI: 10.1124/dmd.114.062604 This article has not been copyedited and formatted. The final version may differ from this version. DMD #62604 Running Title: Differentiation of Human iPS Cells to Enterocytes To whom correspondence should be addressed: Tamihide Matsunaga, Ph.D. Department of Clinical Pharmacy, Graduate School of Pharmaceutical Sciences, Nagoya City University 3-1 Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan Downloaded from Tel, +81-52-836-3751 Fax, +81-52-836-3792 E-mail, [email protected] dmd.aspetjournals.org Text pages: 33 Tables: 1 at ASPET Journals on October 1, 2021 Figures: 7 References: 47 Abstract: 234 words Introduction: 669 words Discussion: 1,051 words 2 DMD Fast Forward.