Using the Novel Dipstick DNA Extraction Technique in a Biological Barcoding Lab
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Louisiana's Animal Species of Greatest Conservation Need (SGCN)
Louisiana's Animal Species of Greatest Conservation Need (SGCN) ‐ Rare, Threatened, and Endangered Animals ‐ 2020 MOLLUSKS Common Name Scientific Name G‐Rank S‐Rank Federal Status State Status Mucket Actinonaias ligamentina G5 S1 Rayed Creekshell Anodontoides radiatus G3 S2 Western Fanshell Cyprogenia aberti G2G3Q SH Butterfly Ellipsaria lineolata G4G5 S1 Elephant‐ear Elliptio crassidens G5 S3 Spike Elliptio dilatata G5 S2S3 Texas Pigtoe Fusconaia askewi G2G3 S3 Ebonyshell Fusconaia ebena G4G5 S3 Round Pearlshell Glebula rotundata G4G5 S4 Pink Mucket Lampsilis abrupta G2 S1 Endangered Endangered Plain Pocketbook Lampsilis cardium G5 S1 Southern Pocketbook Lampsilis ornata G5 S3 Sandbank Pocketbook Lampsilis satura G2 S2 Fatmucket Lampsilis siliquoidea G5 S2 White Heelsplitter Lasmigona complanata G5 S1 Black Sandshell Ligumia recta G4G5 S1 Louisiana Pearlshell Margaritifera hembeli G1 S1 Threatened Threatened Southern Hickorynut Obovaria jacksoniana G2 S1S2 Hickorynut Obovaria olivaria G4 S1 Alabama Hickorynut Obovaria unicolor G3 S1 Mississippi Pigtoe Pleurobema beadleianum G3 S2 Louisiana Pigtoe Pleurobema riddellii G1G2 S1S2 Pyramid Pigtoe Pleurobema rubrum G2G3 S2 Texas Heelsplitter Potamilus amphichaenus G1G2 SH Fat Pocketbook Potamilus capax G2 S1 Endangered Endangered Inflated Heelsplitter Potamilus inflatus G1G2Q S1 Threatened Threatened Ouachita Kidneyshell Ptychobranchus occidentalis G3G4 S1 Rabbitsfoot Quadrula cylindrica G3G4 S1 Threatened Threatened Monkeyface Quadrula metanevra G4 S1 Southern Creekmussel Strophitus subvexus -

Biological Evaluation Usda - Forest Service, Kisatchie National Forest Catahoula Ranger District
Catahoula RD – North Gray Creek 2 Aug 19 BIOLOGICAL EVALUATION USDA - FOREST SERVICE, KISATCHIE NATIONAL FOREST CATAHOULA RANGER DISTRICT North Gray Creek I. INTRODUCTION This report documents the findings of the Biological Evaluation (BE) for the proposed silvicultural activities in Compartments 89-93 on the Catahoula Ranger District of the Kisatchie National Forest. It also serves to provide the decision maker with information and determinations of the effects of proposed actions on proposed, endangered, threatened and sensitive (PETS) species and habitats so that the best decisions can be made regarding these species and the proposal. PETS species are species whose viability is most likely to be put at risk from management actions. Through the BE process the proposed management activities were reviewed and their potential effects on PETS species disclosed. Evaluation methods included internal expertise on species' habitat requirements, field surveys, Forest Service inventory and occurrence records, Final Environmental Impact Statement/Revised Land and Resource Management Plan for the Kisatchie National Forest, the recovery plans for the the red- cockaded woodpecker (RCW) and Louisiana pearlshell mussel (LPM) and the draft recovery plan and candidate conservation agreement (CCA) for the Louisisana pine snake (LPS). This biological evaluation was prepared in accordance with Forest Service Handbook 2609.23R and regulations set forth in Section 7 (a)(2) of the Endangered Species Act. A botanical evaluation was done separately to address impacts on sensitive and conservation plants. PURPOSE AND NEED: Differences between current and desired conditions have been identified within the project area. In order to move the project area toward the desired conditions, specific resource management actions were identified and developed. -

Rare Animals Tracking List
Louisiana's Animal Species of Greatest Conservation Need (SGCN) ‐ Rare, Threatened, and Endangered Animals ‐ 2020 MOLLUSKS Common Name Scientific Name G‐Rank S‐Rank Federal Status State Status Mucket Actinonaias ligamentina G5 S1 Rayed Creekshell Anodontoides radiatus G3 S2 Western Fanshell Cyprogenia aberti G2G3Q SH Butterfly Ellipsaria lineolata G4G5 S1 Elephant‐ear Elliptio crassidens G5 S3 Spike Elliptio dilatata G5 S2S3 Texas Pigtoe Fusconaia askewi G2G3 S3 Ebonyshell Fusconaia ebena G4G5 S3 Round Pearlshell Glebula rotundata G4G5 S4 Pink Mucket Lampsilis abrupta G2 S1 Endangered Endangered Plain Pocketbook Lampsilis cardium G5 S1 Southern Pocketbook Lampsilis ornata G5 S3 Sandbank Pocketbook Lampsilis satura G2 S2 Fatmucket Lampsilis siliquoidea G5 S2 White Heelsplitter Lasmigona complanata G5 S1 Black Sandshell Ligumia recta G4G5 S1 Louisiana Pearlshell Margaritifera hembeli G1 S1 Threatened Threatened Southern Hickorynut Obovaria jacksoniana G2 S1S2 Hickorynut Obovaria olivaria G4 S1 Alabama Hickorynut Obovaria unicolor G3 S1 Mississippi Pigtoe Pleurobema beadleianum G3 S2 Louisiana Pigtoe Pleurobema riddellii G1G2 S1S2 Pyramid Pigtoe Pleurobema rubrum G2G3 S2 Texas Heelsplitter Potamilus amphichaenus G1G2 SH Fat Pocketbook Potamilus capax G2 S1 Endangered Endangered Inflated Heelsplitter Potamilus inflatus G1G2Q S1 Threatened Threatened Ouachita Kidneyshell Ptychobranchus occidentalis G3G4 S1 Rabbitsfoot Quadrula cylindrica G3G4 S1 Threatened Threatened Monkeyface Quadrula metanevra G4 S1 Southern Creekmussel Strophitus subvexus -
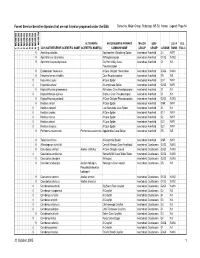
Sensitive Species That Are Not Listed Or Proposed Under the ESA Sorted By: Major Group, Subgroup, NS Sci
Forest Service Sensitive Species that are not listed or proposed under the ESA Sorted by: Major Group, Subgroup, NS Sci. Name; Legend: Page 94 REGION 10 REGION 1 REGION 2 REGION 3 REGION 4 REGION 5 REGION 6 REGION 8 REGION 9 ALTERNATE NATURESERVE PRIMARY MAJOR SUB- U.S. N U.S. 2005 NATURESERVE SCIENTIFIC NAME SCIENTIFIC NAME(S) COMMON NAME GROUP GROUP G RANK RANK ESA C 9 Anahita punctulata Southeastern Wandering Spider Invertebrate Arachnid G4 NNR 9 Apochthonius indianensis A Pseudoscorpion Invertebrate Arachnid G1G2 N1N2 9 Apochthonius paucispinosus Dry Fork Valley Cave Invertebrate Arachnid G1 N1 Pseudoscorpion 9 Erebomaster flavescens A Cave Obligate Harvestman Invertebrate Arachnid G3G4 N3N4 9 Hesperochernes mirabilis Cave Psuedoscorpion Invertebrate Arachnid G5 N5 8 Hypochilus coylei A Cave Spider Invertebrate Arachnid G3? NNR 8 Hypochilus sheari A Lampshade Spider Invertebrate Arachnid G2G3 NNR 9 Kleptochthonius griseomanus An Indiana Cave Pseudoscorpion Invertebrate Arachnid G1 N1 8 Kleptochthonius orpheus Orpheus Cave Pseudoscorpion Invertebrate Arachnid G1 N1 9 Kleptochthonius packardi A Cave Obligate Pseudoscorpion Invertebrate Arachnid G2G3 N2N3 9 Nesticus carteri A Cave Spider Invertebrate Arachnid GNR NNR 8 Nesticus cooperi Lost Nantahala Cave Spider Invertebrate Arachnid G1 N1 8 Nesticus crosbyi A Cave Spider Invertebrate Arachnid G1? NNR 8 Nesticus mimus A Cave Spider Invertebrate Arachnid G2 NNR 8 Nesticus sheari A Cave Spider Invertebrate Arachnid G2? NNR 8 Nesticus silvanus A Cave Spider Invertebrate Arachnid G2? NNR -

BIOLOGICAL EVALUATION (Conservation Species) USDA FOREST SERVICE Kisatchie National Forest Winn Ranger District
BIOLOGICAL EVALUATION (Conservation Species) USDA FOREST SERVICE Kisatchie National Forest Winn Ranger District Mendenhause Project Compartments 51, 52, and 56 INTRODUCTION: The purpose of this Biological Evaluation (BE) is to ensure that Forest Service actions do not contribute to the loss of viability of any native or desirable non-native animal species. This BE documents likely effects of management actions on populations of conservation species of concern as determined by the Louisiana Natural Heritage Program (LNHP). Such species are those, whose viability is most likely to be put at risk from management actions. Information presented here is used to ensure that such species are maintained at, or are moving toward, viable population levels. Populations of other species (those at less risk of losing viability) are maintained by creating and maintaining a diversity of habitat types distributed across the National Forest in accordance with Forest Plan Standards and Guidelines. Through this combination of approaches, viable populations of all species are maintained. A review of the Louisiana Rare Animal Species list and the Forest Geographic Information System (GIS) records was conducted to determine which species would possibly occur within the project site. The general biology of the species, personal communication with experts, and literature searches were used to determine the potential effect proposed actions, including the no action, would have on species considered likely to occur within the project area. The aforementioned research was conducted considering the best available science, and the potential effects discovered are discussed in this BE. PURPOSE AND NEED: Differences between current and desired conditions have been identified within the project area. -

Conservation
CONSERVATION ecapod crustaceans in the families Astacidae, recreational and commercial bait fisheries, and serve as a Cambaridae, and Parastacidae, commonly known profitable and popular food resource. Crayfishes often make as crayfishes or crawfishes, are native inhabitants up a large proportion of the biomass produced in aquatic of freshwater ecosystems on every continent systems (Rabeni 1992; Griffith et al. 1994). In streams, sport except Africa and Antarctica. Although nearly worldwide fishes such as sunfishes and basses (family Centrarchidae) in distribution, crayfishes exhibit the highest diversity in may consume up to two-thirds of the annual production of North America north of Mexico with 338 recognized taxa crayfishes, and as such, crayfishes often comprise critical (308 species and 30 subspecies). Mirroring continental pat- food resources for these fishes (Probst et al. 1984; Roell and terns of freshwater fishes (Warren and Burr 1994) and fresh- Orth 1993). Crayfishes also contribute to the maintenance of water mussels (J. D. Williams et al. 1993), the southeastern food webs by processing vegetation and leaf litter (Huryn United States harbors the highest number of crayfish species. and Wallace 1987; Griffith et al. 1994), which increases avail- Crayfishes are a significant component of aquatic ecosys- ability of nutrients and organic matter to other organisms. tems. They facilitate important ecological processes, sustain In some rivers, bait fisheries for crayfishes constitute an Christopher A. Taylor and Melvin L. Warren, Jr. are cochairs of the Crayfish Subcommittee of the AFS Endangered Species Committee. They can be contacted at the Illinois Natural History Survey, Center for Biodiversity, 607 E. Peabody Drive, Champaign, IL 61820, and U.S. -

Fisheries Conservation Status of Crayfish Species Paddlefish Conservation Case Study
VOL 32 NO 8 AUGUST 2007 Fish News Legislative Update Journal Highlights FisheriesFisheries Calendar American Fisheries Society • www.fisheries.org Job Center Conservation Status of Crayfish Species Paddlefish Conservation Case Study Fisheries • VOL 32 NO 8 • AUGUST 2007 • WWW.FISHERIES.ORG 365 Northwest Marine Tcchnology, Inc. 366 Fisheries • VOL 32 NO 8 • AUGUST 2007 • WWW.FISHERIES.ORG VOL 32 NO 8 AUGUST 2007 372 AMERIFisheriescan FIshERIES SOCIETY • WWW.FIshERIES.ORG EDitOriaL / SUbsCriPtiON / CirCULatiON OffiCES 5410 Grosvenor Lane, Suite 110 • Bethesda, MD 20814-2199 301/897-8616 • fax 301/897-8096 • [email protected] The American Fisheries Society (AFS), founded in 1870, is the oldest and largest professional society representing fisheries scientists. The AFS promotes scientific research and enlightened management of aquatic resources 390 for optimum use and enjoyment by the public. It also XXX encourages comprehensive education of fisheries scientists and continuing on-the-job training. AFS OFFICERS FISHERIES EDITORS Contents STAFF PRESIDENT SENIOR EDITOR SCIENCE Jennifer L. Nielsen Ghassan “Gus” N. EDITORS COLUMN: COLUMN: PRESIDENT ElECT Rassam Madeleine 368 PRESIDENT’S HOOK 398 GUEST DIRECTOR’S LINE Mary C. Fabrizio DIRECTOR OF Hall-Arber New Features for AFS Publications FIRST PUBLICATIONS Ken Ashley Thanks for an Incredible Year VICE PRESIDENT Aaron Lerner Doug Beard As part of an ongoing effort to make AFS William G. Franzin MANAGING Ken Currens Through commitment and hardwork the AFS publications more and more useful for fisheries SECOND EDITOR William E. Kelso volunteer membership has accomplished professionals, several new features have been VICE PRESIDENT Beth Beard Deirdre M. Kimball Donald C. Jackson PRODUCTION Robert T. -

Decapoda, Astacidae)
Vol. 81, pp. 413-418 30 December 1968 PROCEEDINGS OF THE BIOLOGICAL SOCIETY OF WASHINGTON A NEW FAXONELLA FROM NORTHEAST LOUISIANA (DECAPODA, ASTACIDAE) BY JERRY G. WALLS Department of Biological Sciences, McNeese State College, Lake Charles, Louisiana The genus Faxonella (Creaser, 1933) was diagnosed by Fitzpatrick (1963), who recognized two species, F. clypeata (Hay, 1899) and F. beyeri (Penn, 1950). The new species here described brings to three the number of known species. Two of these, F. beyeri and F. creaseri (new species), are endemic to Louisiana. I would like to thank Dr. Joe B. Black, McNeese State College, for the loan of specimens and for his patience and assistance in preparing this description. Thanks are also due to Messrs. Norman Arnold and Frank Amsden for providing transportation and assistance in collecting. Faxonella ereaseri new species Diagnosis: Lateral rostral spines and cervical spines absent; rostrum short and broad, one and one-half or fewer times as long as broad. Shape of chela ( Fig. 4) and carapace as in Faxonella clypeata and F. beyeri; areola variable, 25.9-33.1 percent of carapace length ( average 30.0), and 2.3-5.3 times as long as broad ( average 3.8). Male with copulatory hooks on third pereiopods only. First pleopod of form I male (Figs. 1-3) terminating in two rami: central projection long, flattened, bent mesially; mesial process much shorter, only about one- half as long as central projection, removed from and paralleling central projection. In usual position, terminal elements of left pleopod overlap those of other ( Fig. 3). Annulus ventralis (Fig. -

Chapter 1. Introduction
INTRODUCTION LA WAP—JUNE 2015 CHAPTER 1. INTRODUCTION A. Conservation and Management of Wildlife and Fisheries Resources in Louisiana In Louisiana, the Department of Wildlife and Fisheries (LDWF) is the government agency charged with the conservation and management of wildlife and fishery resources in the state, including aquatic and terrestrial vertebrate and invertebrate species. LDWF is authorized to execute the laws enacted for the control and supervision of programs relating to the management, protection, conservation, and replenishment of wildlife, fish, and aquatic life, and the regulation of the shipping of wildlife, fish, furs, and skins. LDWF is organized into four appropriated budget offices: Secretary, Management and Finance, Wildlife, and Fisheries. 1. Mission Statement: The Louisiana Department of Wildlife and Fisheries (LDWF) is charged with the conservation and management of Louisiana’s natural resources, including both aquatic and terrestrial species and habitats. LDWF’s mission is to manage, conserve, and promote wise utilization of Louisiana’s renewable fish and wildlife resources and their supporting habitats for the social and economic benefit of current and future generations; to provide opportunities for study, utilization, and enjoyment of these resources; and to promote a safe and healthy environment for the users of the resources. B. Problem and Need for a Wildlife Action Plan 1. Background: Early in the twentieth century, many of America’s once numerous fish and wildlife species were on the verge of extinction. In the 1930s, this situation began to change as harvests were better regulated, wildlife management areas and refuges were created, and game species populations were augmented or restored with translocated animals. -

Crustacean Biology Advance Access Published 8 August 2017 Journal of Crustacean Biology the Crustacean Society Journal of Crustacean Biology (2017) 1–39
Journal of Crustacean Biology Advance Access published 8 August 2017 Journal of Crustacean Biology The Crustacean Society Journal of Crustacean Biology (2017) 1–39. doi:10.1093/jcbiol/rux070 An updated classification of the freshwater crayfishes (Decapoda: Astacidea) of the world, with a complete species list Keith A. Crandall1 and Sammy De Grave2 1Computational Biology Institute, George Washington University, SEH 7000D, 800 22nd St. NW, Washington DC, 20052, USA and Department of Invertebrate Zoology, Smithsonian Institution, Washington, DC, USA; and 2Oxford University Museum of Natural History, Parks Road, Oxford OX1 3PW, UK Correspondence: K.A. Crandall; e-mail: [email protected] (Received 13 May 2017; accepted 16 June 2017) ABSTRACT The freshwater crayfishes are a group of decapod crustaceans that have played a critical role in a diversity of biological studies, from physiology, to ecology, neurobiology, conserva- tion, and evolution. Central to many of these fields of study is the dependence on a robust taxonomic framework for accurate communication relating to species diversity and associated attributes. Despite a huge body of taxonomic work since Linnaeus, there has never been a single, comprehensive taxonomic summary of all the species of crayfish of the world. There has also been an abundance of recent taxonomic work in terms of new species descriptions and taxonomic insights gained from a variety of phylogenetic studies. Here we gather diverse taxonomic and phylogenetic information into a single resource. We develop an updated clas- sification system that includes all the crayfishes worldwide and taxonomic changes to better reflect the current phylogenetic knowledge of the group. We also include all the fossil crayfish taxa for a complete classification of extant and extinct crayfishes. -

United States Department of the Interior
United States Department of the Interior FISH AND WILDLIFE SERVICE 200 Dulles Drive Lafayette, Louisiana 70506 August 30, 2019 Ms. Emlyn Smith Wildlife Biologist U.S. Forest Service Kisatchie National Forest Catahoula Ranger District Bentley, LA 71407 Dear Ms. Smith: Please reference your August 7, 2019, email and attachment, which transmits the North Gray Creek biological evaluation (BE) that assesses potential effects of proposed silvicultural activities in Compartments 89-93 on the Catahoula Ranger District of the Kisatchie National Forest in Grant Parish, Louisiana, to the threatened northern long-eared bat (NLEB; Myotis septentrionalis) and threatened Louisiana pearlshell (LPM; Margaritifera hembeli). The Fish and Wildlife Service (Service) has reviewed the information provided and offers the following comments in accordance with the Endangered Species Act (ESA) of 1973 (87 Stat. 884, as amended; 16 U.S.C. 1531 et seq.). As stated in the BE, the purpose to the proposed action is to implement specific resource management activities to move the habitat found within the project area toward the desired future conditions, in accordance with the Revised Land and Resource Management Plan (RLRMP, USFS 1999), with emphasis on maximizing commodity outputs while meeting all minimum management requirements. The proposed action would restore offsite loblolly pine forest to native longleaf and shortleaf pine forest and commercially thin forest stands to enhance health and vigor of the residual forest; to improve species composition, structure, function, and resiliency to insects and disease; and to harvest forest commodity. The proposed action also includes treatments to remove noxious weeds and non-native invasive plants. If implemented, the proposed action would be accomplished within the next five to ten years. -

Population Abundance and Transience of Selected Coastal Plain Crayfishes
Louisiana State University LSU Digital Commons LSU Master's Theses Graduate School 2015 Population Abundance and Transience of Selected Coastal Plain Crayfishes Samantha Lott Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_theses Part of the Environmental Sciences Commons Recommended Citation Lott, Samantha, "Population Abundance and Transience of Selected Coastal Plain Crayfishes" (2015). LSU Master's Theses. 3970. https://digitalcommons.lsu.edu/gradschool_theses/3970 This Thesis is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Master's Theses by an authorized graduate school editor of LSU Digital Commons. For more information, please contact [email protected]. POPULATION ABUNDANCE AND TRANSIENCE OF SELECTED COASTAL PLAIN CRAYFISHES A Thesis Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Master of Science in The School of Renewable Natural Resources by Samantha Lott B.S., The Rochester Institute of Technology, 2012 December 2015 ACKNOWLEDGEMENTS I would like to express my thanks to my fellow graduate students, namely Catherine Reuter and William Budnick, and the undergraduate workers that helped make this project a success. I am particularly grateful for the assistance given by my advisor, Dr. Kaller, who only ever had positive things to say and helped immeasurably with my analytical methods and edits. His willingness to give his time so generously has been much appreciated. ii TABLE OF CONTENTS ACKNOWLEDGEMENTS...........................................................................................................