Model Selection and Diagnostics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Startips …A Resource for Survey Researchers
Subscribe Archives StarTips …a resource for survey researchers Share this article: How to Interpret Standard Deviation and Standard Error in Survey Research Standard Deviation and Standard Error are perhaps the two least understood statistics commonly shown in data tables. The following article is intended to explain their meaning and provide additional insight on how they are used in data analysis. Both statistics are typically shown with the mean of a variable, and in a sense, they both speak about the mean. They are often referred to as the "standard deviation of the mean" and the "standard error of the mean." However, they are not interchangeable and represent very different concepts. Standard Deviation Standard Deviation (often abbreviated as "Std Dev" or "SD") provides an indication of how far the individual responses to a question vary or "deviate" from the mean. SD tells the researcher how spread out the responses are -- are they concentrated around the mean, or scattered far & wide? Did all of your respondents rate your product in the middle of your scale, or did some love it and some hate it? Let's say you've asked respondents to rate your product on a series of attributes on a 5-point scale. The mean for a group of ten respondents (labeled 'A' through 'J' below) for "good value for the money" was 3.2 with a SD of 0.4 and the mean for "product reliability" was 3.4 with a SD of 2.1. At first glance (looking at the means only) it would seem that reliability was rated higher than value. -
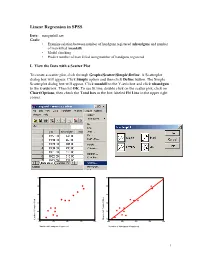
Linear Regression in SPSS
Linear Regression in SPSS Data: mangunkill.sav Goals: • Examine relation between number of handguns registered (nhandgun) and number of man killed (mankill) • Model checking • Predict number of man killed using number of handguns registered I. View the Data with a Scatter Plot To create a scatter plot, click through Graphs\Scatter\Simple\Define. A Scatterplot dialog box will appear. Click Simple option and then click Define button. The Simple Scatterplot dialog box will appear. Click mankill to the Y-axis box and click nhandgun to the x-axis box. Then hit OK. To see fit line, double click on the scatter plot, click on Chart\Options, then check the Total box in the box labeled Fit Line in the upper right corner. 60 60 50 50 40 40 30 30 20 20 10 10 Killed of People Number Number of People Killed 400 500 600 700 800 400 500 600 700 800 Number of Handguns Registered Number of Handguns Registered 1 Click the target button on the left end of the tool bar, the mouse pointer will change shape. Move the target pointer to the data point and click the left mouse button, the case number of the data point will appear on the chart. This will help you to identify the data point in your data sheet. II. Regression Analysis To perform the regression, click on Analyze\Regression\Linear. Place nhandgun in the Dependent box and place mankill in the Independent box. To obtain the 95% confidence interval for the slope, click on the Statistics button at the bottom and then put a check in the box for Confidence Intervals. -

Appendix F.1 SAWG SPC Appendices
Appendix F.1 - SAWG SPC Appendices 8-8-06 Page 1 of 36 Appendix 1: Control Charts for Variables Data – classical Shewhart control chart: When plate counts provide estimates of large levels of organisms, the estimated levels (cfu/ml or cfu/g) can be considered as variables data and the classical control chart procedures can be used. Here it is assumed that the probability of a non-detect is virtually zero. For these types of microbiological data, a log base 10 transformation is used to remove the correlation between means and variances that have been observed often for these types of data and to make the distribution of the output variable used for tracking the process more symmetric than the measured count data1. There are several control charts that may be used to control variables type data. Some of these charts are: the Xi and MR, (Individual and moving range) X and R, (Average and Range), CUSUM, (Cumulative Sum) and X and s, (Average and Standard Deviation). This example includes the Xi and MR charts. The Xi chart just involves plotting the individual results over time. The MR chart involves a slightly more complicated calculation involving taking the difference between the present sample result, Xi and the previous sample result. Xi-1. Thus, the points that are plotted are: MRi = Xi – Xi-1, for values of i = 2, …, n. These charts were chosen to be shown here because they are easy to construct and are common charts used to monitor processes for which control with respect to levels of microbiological organisms is desired. -

Problems with OLS Autocorrelation
Problems with OLS Considering : Yi = α + βXi + ui we assume Eui = 0 2 = σ2 = σ2 E ui or var ui Euiuj = 0orcovui,uj = 0 We have seen that we have to make very specific assumptions about ui in order to get OLS estimates with the desirable properties. If these assumptions don’t hold than the OLS estimators are not necessarily BLU. We can respond to such problems by changing specification and/or changing the method of estimation. First we consider the problems that might occur and what they imply. In all of these we are basically looking at the residuals to see if they are random. ● 1. The errors are serially dependent ⇒ autocorrelation/serial correlation. 2. The error variances are not constant ⇒ heteroscedasticity 3. In multivariate analysis two or more of the independent variables are closely correlated ⇒ multicollinearity 4. The function is non-linear 5. There are problems of outliers or extreme values -but what are outliers? 6. There are problems of missing variables ⇒can lead to missing variable bias Of course these problems do not have to come separately, nor are they likely to ● Note that in terms of significance things may look OK and even the R2the regression may not look that bad. ● Really want to be able to identify a misleading regression that you may take seriously when you should not. ● The tests in Microfit cover many of the above concerns, but you should always plot the residuals and look at them. Autocorrelation This implies that taking the time series regression Yt = α + βXt + ut but in this case there is some relation between the error terms across observations. -
Model Selection for Optimal Prediction in Statistical Machine Learning
Model Selection for Optimal Prediction in Statistical Machine Learning Ernest Fokou´e Introduction science with several ideas from cognitive neuroscience and At the core of all our modern-day advances in artificial in- psychology to inspire the creation, invention, and discov- telligence is the emerging field of statistical machine learn- ery of abstract models that attempt to learn and extract pat- ing (SML). From a very general perspective, SML can be terns from the data. One could think of SML as a field thought of as a field of mathematical sciences that com- of science dedicated to building models endowed with bines mathematics, probability, statistics, and computer the ability to learn from the data in ways similar to the ways humans learn, with the ultimate goal of understand- Ernest Fokou´eis a professor of statistics at Rochester Institute of Technology. His ing and then mastering our complex world well enough email address is [email protected]. to predict its unfolding as accurately as possible. One Communicated by Notices Associate Editor Emilie Purvine. of the earliest applications of statistical machine learning For permission to reprint this article, please contact: centered around the now ubiquitous MNIST benchmark [email protected]. task, which consists of building statistical models (also DOI: https://doi.org/10.1090/noti2014 FEBRUARY 2020 NOTICES OF THE AMERICAN MATHEMATICAL SOCIETY 155 known as learning machines) that automatically learn and Theoretical Foundations accurately recognize handwritten digits from the United It is typical in statistical machine learning that a given States Postal Service (USPS). A typical deployment of an problem will be solved in a wide variety of different ways. -

The Bootstrap and Jackknife
The Bootstrap and Jackknife Summer 2017 Summer Institutes 249 Bootstrap & Jackknife Motivation In scientific research • Interest often focuses upon the estimation of some unknown parameter, θ. The parameter θ can represent for example, mean weight of a certain strain of mice, heritability index, a genetic component of variation, a mutation rate, etc. • Two key questions need to be addressed: 1. How do we estimate θ ? 2. Given an estimator for θ , how do we estimate its precision/accuracy? • We assume Question 1 can be reasonably well specified by the researcher • Question 2, for our purposes, will be addressed via the estimation of the estimator’s standard error Summer 2017 Summer Institutes 250 What is a standard error? Suppose we want to estimate a parameter theta (eg. the mean/median/squared-log-mode) of a distribution • Our sample is random, so… • Any function of our sample is random, so... • Our estimate, theta-hat, is random! So... • If we collected a new sample, we’d get a new estimate. Same for another sample, and another... So • Our estimate has a distribution! It’s called a sampling distribution! The standard deviation of that distribution is the standard error Summer 2017 Summer Institutes 251 Bootstrap Motivation Challenges • Answering Question 2, even for relatively simple estimators (e.g., ratios and other non-linear functions of estimators) can be quite challenging • Solutions to most estimators are mathematically intractable or too complicated to develop (with or without advanced training in statistical inference) • However • Great strides in computing, particularly in the last 25 years, have made computational intensive calculations feasible. -

Lecture 5 Significance Tests Criticisms of the NHST Publication Bias Research Planning
Lecture 5 Significance tests Criticisms of the NHST Publication bias Research planning Theophanis Tsandilas !1 Calculating p The p value is the probability of obtaining a statistic as extreme or more extreme than the one observed if the null hypothesis was true. When data are sampled from a known distribution, an exact p can be calculated. If the distribution is unknown, it may be possible to estimate p. 2 Normal distributions If the sampling distribution of the statistic is normal, we will use the standard normal distribution z to derive the p value 3 Example An experiment studies the IQ scores of people lacking enough sleep. H0: μ = 100 and H1: μ < 100 (one-sided) or H0: μ = 100 and H1: μ = 100 (two-sided) 6 4 Example Results from a sample of 15 participants are as follows: 90, 91, 93, 100, 101, 88, 98, 100, 87, 83, 97, 105, 99, 91, 81 The mean IQ score of the above sample is M = 93.6. Is this value statistically significantly different than 100? 5 Creating the test statistic We assume that the population standard deviation is known and equal to SD = 15. Then, the standard error of the mean is: σ 15 σµˆ = = =3.88 pn p15 6 Creating the test statistic We assume that the population standard deviation is known and equal to SD = 15. Then, the standard error of the mean is: σ 15 σµˆ = = =3.88 pn p15 The test statistic tests the standardized difference between the observed mean µ ˆ = 93 . 6 and µ 0 = 100 µˆ µ 93.6 100 z = − 0 = − = 1.65 σµˆ 3.88 − The p value is the probability of getting a z statistic as or more extreme than this value (given that H0 is true) 7 Calculating the p value µˆ µ 93.6 100 z = − 0 = − = 1.65 σµˆ 3.88 − The p value is the probability of getting a z statistic as or more extreme than this value (given that H0 is true) 8 Calculating the p value To calculate the area in the distribution, we will work with the cumulative density probability function (cdf). -

Linear Regression: Goodness of Fit and Model Selection
Linear Regression: Goodness of Fit and Model Selection 1 Goodness of Fit I Goodness of fit measures for linear regression are attempts to understand how well a model fits a given set of data. I Models almost never describe the process that generated a dataset exactly I Models approximate reality I However, even models that approximate reality can be used to draw useful inferences or to prediction future observations I ’All Models are wrong, but some are useful’ - George Box 2 Goodness of Fit I We have seen how to check the modelling assumptions of linear regression: I checking the linearity assumption I checking for outliers I checking the normality assumption I checking the distribution of the residuals does not depend on the predictors I These are essential qualitative checks of goodness of fit 3 Sample Size I When making visual checks of data for goodness of fit is important to consider sample size I From a multiple regression model with 2 predictors: I On the left is a histogram of the residuals I On the right is residual vs predictor plot for each of the two predictors 4 Sample Size I The histogram doesn’t look normal but there are only 20 datapoint I We should not expect a better visual fit I Inferences from the linear model should be valid 5 Outliers I Often (particularly when a large dataset is large): I the majority of the residuals will satisfy the model checking assumption I a small number of residuals will violate the normality assumption: they will be very big or very small I Outliers are often generated by a process distinct from those which we are primarily interested in. -

Probability Distributions and Error Bars
Statistics and Data Analysis in MATLAB Kendrick Kay, [email protected] Lecture 1: Probability distributions and error bars 1. Exploring a simple dataset: one variable, one condition - Let's start with the simplest possible dataset. Suppose we measure a single quantity for a single condition. For example, suppose we measure the heights of male adults. What can we do with the data? - The histogram provides a useful summary of a set of data—it shows the distribution of the data. A histogram is constructed by binning values and counting the number of observations in each bin. - The mean and standard deviation are simple summaries of a set of data. They are parametric statistics, as they make implicit assumptions about the form of the data. The mean is designed to quantify the central tendency of a set of data, while the standard deviation is designed to quantify the spread of a set of data. n ∑ xi mean(x) = x = i=1 n n 2 ∑(xi − x) std(x) = i=1 n − 1 In these equations, xi is the ith data point and n is the total number of data points. - The median and interquartile range (IQR) also summarize data. They are nonparametric statistics, as they make minimal assumptions about the form of the data. The Xth percentile is the value below which X% of the data points lie. The median is the 50th percentile. The IQR is the difference between the 75th and 25th percentiles. - Mean and standard deviation are appropriate when the data are roughly Gaussian. When the data are not Gaussian (e.g. -

Scalable Model Selection for Spatial Additive Mixed Modeling: Application to Crime Analysis
Scalable model selection for spatial additive mixed modeling: application to crime analysis Daisuke Murakami1,2,*, Mami Kajita1, Seiji Kajita1 1Singular Perturbations Co. Ltd., 1-5-6 Risona Kudan Building, Kudanshita, Chiyoda, Tokyo, 102-0074, Japan 2Department of Statistical Data Science, Institute of Statistical Mathematics, 10-3 Midori-cho, Tachikawa, Tokyo, 190-8562, Japan * Corresponding author (Email: [email protected]) Abstract: A rapid growth in spatial open datasets has led to a huge demand for regression approaches accommodating spatial and non-spatial effects in big data. Regression model selection is particularly important to stably estimate flexible regression models. However, conventional methods can be slow for large samples. Hence, we develop a fast and practical model-selection approach for spatial regression models, focusing on the selection of coefficient types that include constant, spatially varying, and non-spatially varying coefficients. A pre-processing approach, which replaces data matrices with small inner products through dimension reduction dramatically accelerates the computation speed of model selection. Numerical experiments show that our approach selects the model accurately and computationally efficiently, highlighting the importance of model selection in the spatial regression context. Then, the present approach is applied to open data to investigate local factors affecting crime in Japan. The results suggest that our approach is useful not only for selecting factors influencing crime risk but also for predicting crime events. This scalable model selection will be key to appropriately specifying flexible and large-scale spatial regression models in the era of big data. The developed model selection approach was implemented in the R package spmoran. Keywords: model selection; spatial regression; crime; fast computation; spatially varying coefficient modeling 1. -

A Robust Rescaled Moment Test for Normality in Regression
Journal of Mathematics and Statistics 5 (1): 54-62, 2009 ISSN 1549-3644 © 2009 Science Publications A Robust Rescaled Moment Test for Normality in Regression 1Md.Sohel Rana, 1Habshah Midi and 2A.H.M. Rahmatullah Imon 1Laboratory of Applied and Computational Statistics, Institute for Mathematical Research, University Putra Malaysia, 43400 Serdang, Selangor, Malaysia 2Department of Mathematical Sciences, Ball State University, Muncie, IN 47306, USA Abstract: Problem statement: Most of the statistical procedures heavily depend on normality assumption of observations. In regression, we assumed that the random disturbances were normally distributed. Since the disturbances were unobserved, normality tests were done on regression residuals. But it is now evident that normality tests on residuals suffer from superimposed normality and often possess very poor power. Approach: This study showed that normality tests suffer huge set back in the presence of outliers. We proposed a new robust omnibus test based on rescaled moments and coefficients of skewness and kurtosis of residuals that we call robust rescaled moment test. Results: Numerical examples and Monte Carlo simulations showed that this proposed test performs better than the existing tests for normality in the presence of outliers. Conclusion/Recommendation: We recommend using our proposed omnibus test instead of the existing tests for checking the normality of the regression residuals. Key words: Regression residuals, outlier, rescaled moments, skewness, kurtosis, jarque-bera test, robust rescaled moment test INTRODUCTION out that the powers of t and F tests are extremely sensitive to the hypothesized error distribution and may In regression analysis, it is a common practice over deteriorate very rapidly as the error distribution the years to use the Ordinary Least Squares (OLS) becomes long-tailed. -

Model Selection Techniques: an Overview
Model Selection Techniques An overview ©ISTOCKPHOTO.COM/GREMLIN Jie Ding, Vahid Tarokh, and Yuhong Yang n the era of big data, analysts usually explore various statis- following different philosophies and exhibiting varying per- tical models or machine-learning methods for observed formances. The purpose of this article is to provide a compre- data to facilitate scientific discoveries or gain predictive hensive overview of them, in terms of their motivation, large power. Whatever data and fitting procedures are employed, sample performance, and applicability. We provide integrated Ia crucial step is to select the most appropriate model or meth- and practically relevant discussions on theoretical properties od from a set of candidates. Model selection is a key ingredi- of state-of-the-art model selection approaches. We also share ent in data analysis for reliable and reproducible statistical our thoughts on some controversial views on the practice of inference or prediction, and thus it is central to scientific stud- model selection. ies in such fields as ecology, economics, engineering, finance, political science, biology, and epidemiology. There has been a Why model selection long history of model selection techniques that arise from Vast developments in hardware storage, precision instrument researches in statistics, information theory, and signal process- manufacturing, economic globalization, and so forth have ing. A considerable number of methods has been proposed, generated huge volumes of data that can be analyzed to extract useful information. Typical statistical inference or machine- learning procedures learn from and make predictions on data Digital Object Identifier 10.1109/MSP.2018.2867638 Date of publication: 13 November 2018 by fitting parametric or nonparametric models (in a broad 16 IEEE SIGNAL PROCESSING MAGAZINE | November 2018 | 1053-5888/18©2018IEEE sense).