Improving Python with Fortran
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Xcode Package from App Store
KH Computational Physics- 2016 Introduction Setting up your computing environment Installation • MAC or Linux are the preferred operating system in this course on scientific computing. • Windows can be used, but the most important programs must be installed – python : There is a nice package ”Enthought Python Distribution” http://www.enthought.com/products/edudownload.php – C++ and Fortran compiler – BLAS&LAPACK for linear algebra – plotting program such as gnuplot Kristjan Haule, 2016 –1– KH Computational Physics- 2016 Introduction Software for this course: Essentials: • Python, and its packages in particular numpy, scipy, matplotlib • C++ compiler such as gcc • Text editor for coding (for example Emacs, Aquamacs, Enthought’s IDLE) • make to execute makefiles Highly Recommended: • Fortran compiler, such as gfortran or intel fortran • BLAS& LAPACK library for linear algebra (most likely provided by vendor) • open mp enabled fortran and C++ compiler Useful: • gnuplot for fast plotting. • gsl (Gnu scientific library) for implementation of various scientific algorithms. Kristjan Haule, 2016 –2– KH Computational Physics- 2016 Introduction Installation on MAC • Install Xcode package from App Store. • Install ‘‘Command Line Tools’’ from Apple’s software site. For Mavericks and lafter, open Xcode program, and choose from the menu Xcode -> Open Developer Tool -> More Developer Tools... You will be linked to the Apple page that allows you to access downloads for Xcode. You wil have to register as a developer (free). Search for the Xcode Command Line Tools in the search box in the upper left. Download and install the correct version of the Command Line Tools, for example for OS ”El Capitan” and Xcode 7.2, Kristjan Haule, 2016 –3– KH Computational Physics- 2016 Introduction you need Command Line Tools OS X 10.11 for Xcode 7.2 Apple’s Xcode contains many libraries and compilers for Mac systems. -

Red Hat Developer Toolset 9 User Guide
Red Hat Developer Toolset 9 User Guide Installing and Using Red Hat Developer Toolset Last Updated: 2020-08-07 Red Hat Developer Toolset 9 User Guide Installing and Using Red Hat Developer Toolset Zuzana Zoubková Red Hat Customer Content Services Olga Tikhomirova Red Hat Customer Content Services [email protected] Supriya Takkhi Red Hat Customer Content Services Jaromír Hradílek Red Hat Customer Content Services Matt Newsome Red Hat Software Engineering Robert Krátký Red Hat Customer Content Services Vladimír Slávik Red Hat Customer Content Services Legal Notice Copyright © 2020 Red Hat, Inc. The text of and illustrations in this document are licensed by Red Hat under a Creative Commons Attribution–Share Alike 3.0 Unported license ("CC-BY-SA"). An explanation of CC-BY-SA is available at http://creativecommons.org/licenses/by-sa/3.0/ . In accordance with CC-BY-SA, if you distribute this document or an adaptation of it, you must provide the URL for the original version. Red Hat, as the licensor of this document, waives the right to enforce, and agrees not to assert, Section 4d of CC-BY-SA to the fullest extent permitted by applicable law. Red Hat, Red Hat Enterprise Linux, the Shadowman logo, the Red Hat logo, JBoss, OpenShift, Fedora, the Infinity logo, and RHCE are trademarks of Red Hat, Inc., registered in the United States and other countries. Linux ® is the registered trademark of Linus Torvalds in the United States and other countries. Java ® is a registered trademark of Oracle and/or its affiliates. XFS ® is a trademark of Silicon Graphics International Corp. -

Download Chapter 3: "Configuring Your Project with Autoconf"
CONFIGURING YOUR PROJECT WITH AUTOCONF Come my friends, ’Tis not too late to seek a newer world. —Alfred, Lord Tennyson, “Ulysses” Because Automake and Libtool are essen- tially add-on components to the original Autoconf framework, it’s useful to spend some time focusing on using Autoconf without Automake and Libtool. This will provide a fair amount of insight into how Autoconf operates by exposing aspects of the tool that are often hidden by Automake. Before Automake came along, Autoconf was used alone. In fact, many legacy open source projects never made the transition from Autoconf to the full GNU Autotools suite. As a result, it’s not unusual to find a file called configure.in (the original Autoconf naming convention) as well as handwritten Makefile.in templates in older open source projects. In this chapter, I’ll show you how to add an Autoconf build system to an existing project. I’ll spend most of this chapter talking about the fundamental features of Autoconf, and in Chapter 4, I’ll go into much more detail about how some of the more complex Autoconf macros work and how to properly use them. Throughout this process, we’ll continue using the Jupiter project as our example. Autotools © 2010 by John Calcote Autoconf Configuration Scripts The input to the autoconf program is shell script sprinkled with macro calls. The input stream must also include the definitions of all referenced macros—both those that Autoconf provides and those that you write yourself. The macro language used in Autoconf is called M4. (The name means M, plus 4 more letters, or the word Macro.1) The m4 utility is a general-purpose macro language processor originally written by Brian Kernighan and Dennis Ritchie in 1977. -
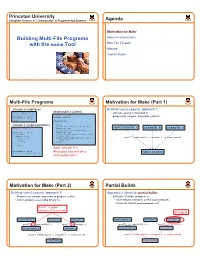
Building Multi-File Programs with the Make Tool
Princeton University Computer Science 217: Introduction to Programming Systems Agenda Motivation for Make Building Multi-File Programs Make Fundamentals with the make Tool Non-File Targets Macros Implicit Rules 1 2 Multi-File Programs Motivation for Make (Part 1) intmath.h (interface) Building testintmath, approach 1: testintmath.c (client) #ifndef INTMATH_INCLUDED • Use one gcc217 command to #define INTMATH_INCLUDED #include "intmath.h" int gcd(int i, int j); #include <stdio.h> preprocess, compile, assemble, and link int lcm(int i, int j); #endif int main(void) { int i; intmath.c (implementation) int j; printf("Enter the first integer:\n"); #include "intmath.h" scanf("%d", &i); testintmath.c intmath.h intmath.c printf("Enter the second integer:\n"); int gcd(int i, int j) scanf("%d", &j); { int temp; printf("Greatest common divisor: %d.\n", while (j != 0) gcd(i, j)); { temp = i % j; printf("Least common multiple: %d.\n", gcc217 testintmath.c intmath.c –o testintmath i = j; lcm(i, j); j = temp; return 0; } } return i; } Note: intmath.h is int lcm(int i, int j) #included into intmath.c { return (i / gcd(i, j)) * j; testintmath } and testintmath.c 3 4 Motivation for Make (Part 2) Partial Builds Building testintmath, approach 2: Approach 2 allows for partial builds • Preprocess, compile, assemble to produce .o files • Example: Change intmath.c • Link to produce executable binary file • Must rebuild intmath.o and testintmath • Need not rebuild testintmath.o!!! Recall: -c option tells gcc217 to omit link changed testintmath.c intmath.h intmath.c -
Failed to Download the Gnu Archive Emacs 'Failed to Download `Marmalade' Archive', but I See the List in Wireshark
failed to download the gnu archive emacs 'Failed to download `marmalade' archive', but I see the list in Wireshark. I've got 129 packets from marmalade-repo.org , many of which list Marmalade package entries, in my Wireshark log. I'm not behind a proxy and HTTP_PROXY is unset. And ELPA (at 'http://tromey.com/elpa/') works fine. I'm on Max OS X Mavericks, all-up-to-date, with Aquamacs, and using the package.el (byte-compiled) as described here: http://marmalade- repo.org/ (since I am on < Emacs 24). What are the next troubleshooting steps I should take? 1 Answer 1. I've taken Aaron Miller's suggestion and fully migrated to the OS X port of Emacs 24.3 . I do miss being able to use the 'command' key to go to the top of the current file, and the slightly smoother gui of Aquamacs, but it's no doubt a great port. Due to the issue with Marmalade, Emacs 23.4 won't work with some of the packages I now need (unless they were hand-built). Installing ELPA Package System for Emacs 23. This page is a guide on installing ELPA package system package.el for emacs 23. If you are using emacs 24, see: Emacs: Install Package with ELPA/MELPA. (type Alt + x version or in terminal emacs --version .) To install, paste the following in a empty file: then call eval-buffer . That will download the file and place it at. (very neat elisp trick! note to self: url-retrieve-synchronously is bundled with emacs 23. -

Ledger Mode Emacs Support for Version 3.0 of Ledger
Ledger Mode Emacs Support For Version 3.0 of Ledger Craig Earls Copyright c 2013, Craig Earls. All rights reserved. Redistribution and use in source and binary forms, with or without modification, are per- mitted provided that the following conditions are met: • Redistributions of source code must retain the above copyright notice, this list of con- ditions and the following disclaimer. • Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. • Neither the name of New Artisans LLC nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBU- TORS \AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAM- AGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTER- RUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. i Table of Contents -

Latex on Macintosh
LaTeX on Macintosh Installing MacTeX and using TeXShop, as described on the main LaTeX page, is enough to get you started using LaTeX on a Mac. This page provides further information for experienced users. Tips for using TeXShop Forward and Inverse Search If you are working on a long document, forward and inverse searching make editing much easier. • Forward search means jumping from a point in your LaTeX source file to the corresponding line in the pdf output file. • Inverse search means jumping from a line in the pdf file back to the corresponding point in the source file. In TeXShop, forward and inverse search are easy. To do a forward search, right-click on text in the source window and choose the option "Sync". (Shortcut: Cmd-Click). Similarly, to do an inverse search, right-click in the output window and choose "Sync". Spell-checking TeXShop does spell-checking using Apple's built in dictionaries, but the results aren't ideal, since TeXShop will mark most LaTeX commands as misspellings, and this will distract you from the real misspellings in your document. Below are two ways around this problem. • Use the spell-checking program Excalibur which is included with MacTeX (in the folder / Applications/TeX). Before you start spell-checking with Excalibur, open its Preferences, and in the LaTeX section, turn on "LaTeX command parsing". Then, Excalibur will ignore LaTeX commands when spell-checking. • Install CocoAspell (already installed in the Math Lab), then go to System Preferences, and open the Spelling preference pane. Choose the last dictionary in the list (the third one labeled "English (United States)"), and check the box for "TeX/LaTeX filtering". -

Installation of R, of R Packages, and Editor Environments
Installation of R, of R packages, and Editor Environments John Maindonald July 11, 2013 Most users (Windows, MacOS X and some flavours of Linux) will be able to install R from binaries. Download the appropriate file, place it in a convenient folder (perhaps the desktop), click on the file icon. If in doubt, accept the defaults for the prompts that appear. Australian users should get the binaries (or source files, if needed) from an Australian Comprehensive R Archive Network (CRAN) mirror site: http://cran.csiro.au or http://cran.ms.unimelb.edu.au/. For installation under MaxOS X, see later. 1 R under Windows 1.1 The Startup Working Directory If left at the installation default, the startup directory for a user who is not logged in as administrator, is likely to be: "C:nDocuments and SettingsnOwnernMy Documents". This can be changed to any directory that has write access, or new icons created with new startup directories. A good strategy is to have a separate working directory for each new project. In the image below C:nr-course is used, but any directory that has write access is possible. To create an icon that starts in a new working directory, make a copy of the R icon. Then right click on the copy and set the working directory (under Target, in the Shortcut tab of Properties) to the directory that you intend to use for the course. 1.2 Installation of Packages Most users will want to install R packages that are additional to those included in the initial installation. -

CSE341: Programming Languages Using SML and Emacs Contents 1
CSE341: Programming Languages Using SML and Emacs Autumn 2018 Contents 1 Overview ................................................. 1 2 Using the Department Labs ...................................... 1 3 Emacs Installation ............................................ 2 4 Emacs Basics ............................................... 2 5 SML/NJ Installation .......................................... 5 6 SML Mode for Emacs Installation ................................. 6 7 Using the SML/NJ REPL (Read-Eval-Print Loop) in Emacs ................ 7 1 Overview For roughly the first half of the course, we will work with the Standard ML programming language, using the SML/NJ (Standard ML of New Jersey) compiler. You will need SML/NJ and a text editor on your computer to do the programming assignments. Any editor that can handle plain text will work, but we recommend using Emacs and its SML Mode, which provides good syntax highlighting, indentation, and integration with an SML environment. While Emacs does not have the look-and-feel or tool-integration of many modern integrated development environments (IDEs), it is a versatile tool well-known by many computer scientists and software developers. This document describes how to install, configure, and use Emacs, SML/NJ, and SML-Mode-for-emacs (henceforth SML Mode) on your computer. These instructions should work for recent versions of Windows, Mac OS X, and Linux. We also describe how to use the machines in the CSE undergraduate Allen School labs or virtual machines, where most of what you need is already installed on both the Windows and Linux machines. 2 Using the Department Labs If you use the machines in the basement labs or the Allen School virtual machines (https://www.cs. washington.edu/lab/software/linuxhomevm), then recent versions of Emacs and SML/NJ should already be installed for you, allowing you to skip Sections 3 and 5 below. -

Assessing Unikernel Security April 2, 2019 – Version 1.0
NCC Group Whitepaper Assessing Unikernel Security April 2, 2019 – Version 1.0 Prepared by Spencer Michaels Jeff Dileo Abstract Unikernels are small, specialized, single-address-space machine images constructed by treating component applications and drivers like libraries and compiling them, along with a kernel and a thin OS layer, into a single binary blob. Proponents of unikernels claim that their smaller codebase and lack of excess services make them more efficient and secure than full-OS virtual machines and containers. We surveyed two major unikernels, Rumprun and IncludeOS, and found that this was decidedly not the case: unikernels, which in many ways resemble embedded systems, appear to have a similarly minimal level of security. Features like ASLR, W^X, stack canaries, heap integrity checks and more are either completely absent or seriously flawed. If an application running on such a system contains a memory corruption vulnerability, it is often possible for attackers to gain code execution, even in cases where the applica- tion’s source and binary are unknown. Furthermore, because the application and the kernel run together as a single process, an attacker who compromises a unikernel can immediately exploit functionality that would require privilege escalation on a regular OS, e.g. arbitrary packet I/O. We demonstrate such attacks on both Rumprun and IncludeOS unikernels, and recommend measures to mitigate them. Table of Contents 1 Introduction to Unikernels . 4 2 Threat Model Considerations . 5 2.1 Unikernel Capabilities ................................................................ 5 2.2 Unikernels Versus Containers .......................................................... 5 3 Hypothesis . 6 4 Testing Methodology . 7 4.1 ASLR ................................................................................ 7 4.2 Page Protections .................................................................... -

AMD Optimizing CPU Libraries User Guide Version 2.1
[AMD Public Use] AMD Optimizing CPU Libraries User Guide Version 2.1 [AMD Public Use] AOCL User Guide 2.1 Table of Contents 1. Introduction .......................................................................................................................................... 4 2. Supported Operating Systems and Compliers ...................................................................................... 5 3. BLIS library for AMD .............................................................................................................................. 6 3.1. Installation ........................................................................................................................................ 6 3.1.1. Build BLIS from source .................................................................................................................. 6 3.1.1.1. Single-thread BLIS ..................................................................................................................... 6 3.1.1.2. Multi-threaded BLIS .................................................................................................................. 6 3.1.2. Using pre-built binaries ................................................................................................................. 7 3.2. Usage ................................................................................................................................................. 7 3.2.1. BLIS Usage in FORTRAN ................................................................................................................ -

Evaluating Elisp Code
Contents Contents Introduction Thank You .................... Intended Audience ................ What You’ll Learn . The Way of Emacs Guiding Philosophy . LISP? ..................... Extensibility . Important Conventions . The Buffer . The Window and the Frame . The Point and Mark . Killing, Yanking and CUA . .emacs.d, init.el, and .emacs . Major Modes and Minor Modes . First Steps Installing and Starting Emacs . Starting Emacs . The Emacs Interface . Keys ........................ Caps Lock as Control . M-x: Execute Extended Command . Universal Arguments . Discovering and Remembering Keys . Configuring Emacs . The Customize Interface . Evaluating Elisp Code . The Package Manager . Color Themes . Getting Help ................... The Info Manual . Apropos ................... The Describe System . The Theory of Movement The Basics ..................... C-x C-f: Find file . C-x C-s: Save Buffer . C-x C-c: Exits Emacs . C-x b: Switch Buffer . C-x k: Kill Buffer . ESC ESC ESC: Keyboard Escape . C-/: Undo . Window Management . Working with Other Windows . Frame Management . Elemental Movement . Navigation Keys . Moving by Character . Moving by Line . Moving by Word . Moving by S-Expressions . Other Movement Commands . Scrolling . Bookmarks and Registers . Selections and Regions . Selection Compatibility Modes . Setting the Mark . Searching and Indexing . Isearch: Incremental Search . Occur: Print lines matching an expression . Imenu: Jump to definitions . Helm: Incremental Completion and Selection IDO: Interactively DO Things . Grep: Searching the file system . Other Movement Commands . Conclusion . The Theory of Editing Killing and Yanking Text . Killing versus Deleting . Yanking Text . Transposing Text . C-t: Transpose Characters . M-t: Transpose Words . C-M-t: Transpose S-expressions . Other Transpose Commands . Filling and Commenting . Filling . Commenting . Search and Replace . Case Folding . Regular Expressions . Changing Case . Counting Things .