Transcriptional Correlates of Proximal-Distal Identify and Regeneration Timing in Axolotl Limbs S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

A Novel Keratin 12 Mutation in a German Kindred with Meesmann's
Br J Ophthalmol 2000;84:527–530 527 A novel keratin 12 mutation in a German kindred Br J Ophthalmol: first published as 10.1136/bjo.84.5.527 on 1 May 2000. Downloaded from with Meesmann’s corneal dystrophy L D Corden, O Swensson, B Swensson, R Rochels, B Wannke, H J Thiel, W H I McLean Abstract inherited disorder aVecting the corneal epithe- Aim—To study a kindred with Mees- lium of both eyes.1–3 It manifests in early child- mann’s corneal dystrophy (MCD) to de- hood and is characterised by myriads of termine if a mutation within the cornea intraepithelial microcysts of variable distribu- specific K3 or K12 genes is responsible for tion and density. The microcysts are best seen the disease phenotype. by retroillumination and slit lamp Methods—Slit lamp examination of the examination.45 Further clinical signs of the cornea in four members of the kindred disease are recurrent punctate erosions with lacrimation and photophobia. Symptoms in- Epithelial Genetics was carried out to confirm the diagnosis Group, Human of MCD. The region encoding the helix clude blurred vision and foreign body sensa- Genetics Unit, initiation motif (HIM) of the K12 polypep- tion. Histologically, the corneal epithelium Department of tide was polymerase chain reaction (PCR) appears irregularly thickened and contains Molecular and Cellular amplified from genomic DNA derived numerous vacuolated suprabasal cells.6–8 In Pathology, Ninewells addition, there is intraepithelial cyst formation Medical School, from aVected individuals in the kindred. Dundee DD1 9SY PCR products generated were subjected to with accumulated periodic acid SchiV (PAS) L D Corden direct automated sequencing. -

Keratin 12-Deficient Mice Have Fragile Corneal Epithelia
Keratin 12-Deficient Mice Have Fragile Corneal Epithelia Winston W.—Y. Kao,* Chia-YangLiu,* Richard L. Converse,* Atsushi Shiraishi* Candace W.-C. Kao,* Masamichi Ishizaki* Thomas Doetschman,^ and John Duffy-f Purpose. Expression of the K3-K12 keratin pair characterizes the corneal epithelial differentia- tion. To elucidate the role of keratin 12 in the maintenance of corneal epithelium integrity, the authors bred mice deficient in keratin 12 by gene-targeting techniques. Methods. One allele of murine Krtl.12 gene was ablated in the embryonic stem cell line, E14.1, by homologous recombination with a DNA construct in which the DNA element between intron 2 and exon 8 of the keratin 12 gene was replaced by a neo-gene. The homologous recombinant embryonic stem cells were injected to mouse blastocysts, and germ lines of chimeras were obtained. The corneas of heterozygous and homozygous mice were characterized by clinical observations using stereomicroscopy, histology with light and electron microscopy, Western immunoblot analysis, immunohistochemistry, in situ hybridization, and Northern hybridization. Results. The heterozygous mice (+/—) one allele of the Krtl.12 gene appear normal and do not develop any clinical manifestations (e.g., corneal epithelial defects). Homozygous mice (—/—) develop normally and suffer mild corneal epithelial erosion. Their corneal epithelia are fragile and can be removed by gentle rubbing of the eyes or brushing with a Microsponge. The corneal epithelium of the homozygote (—/—) does not express keratin 12 as judged by immunohistochemis- try, Western immunoblot analysis with epitope-specific anti-keratin 12 antibodies, Nordiern hybrid- ization with 32P-labeled keratin 12 cDNA, and in situ hybridization with an anti-sense keratin 12 riboprobe. -

S Epithelial Corneal Dystrophy Phenotype Due to A
Eye (2013) 27, 367–373 & 2013 Macmillan Publishers Limited All rights reserved 0950-222X/13 www.nature.com/eye 1 2 2 2 Severe Meesmann’s H Hassan , C Thaung , ND Ebenezer , G Larkin , CLINICAL STUDY AJ Hardcastle2 and SJ Tuft1;2 epithelial corneal dystrophy phenotype due to a missense mutation in the helix- initiation motif of keratin 12 Abstract Purpose To describe a severe phenotype of protein. The clinical effects are markedly Meesmann’s epithelial corneal dystrophy more severe than the phenotype usually (MECD) and to determine the underlying associated with the Arg135Thr mutation molecular cause. within this motif, most frequently seen in Methods We identified a 30-member family European patients with MECD. affected by MECD and examined 11 of the 14 Eye (2013) 27, 367–373; doi:10.1038/eye.2012.261; affected individuals. Excised corneal tissue published online 7 December 2012 from one affected individual was examined histologically. We used PCR and direct Keywords: corneal epithelium; Meesmann sequencing to identify mutation of the corneal dystrophy; human KRT12 protein coding regions of the KRT3 and KRT12 genes. Introduction Results Cases had an unusually severe phenotype with large numbers of Meesmann’s epithelial corneal dystrophy 1Corneal Service, Moorfields intraepithelial cysts present from infancy and (MECD; OMIM #122100) is an autosomal Eye Hospital NHS they developed subepithelial fibrosis in the dominant inherited change of the corneal Foundation Trust, London, second to third decade. In some individuals, epithelium of the eye characterized by the UK the cornea became superficially vascularized, bilaterally symmetric development of numerous 2 a change accompanied by the loss of intraepithelial cysts.1,2 The cysts are best Department of Ocular clinically obvious epithelial cysts. -

Genomics of Inherited Bone Marrow Failure and Myelodysplasia Michael
Genomics of inherited bone marrow failure and myelodysplasia Michael Yu Zhang A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2015 Reading Committee: Mary-Claire King, Chair Akiko Shimamura Marshall Horwitz Program Authorized to Offer Degree: Molecular and Cellular Biology 1 ©Copyright 2015 Michael Yu Zhang 2 University of Washington ABSTRACT Genomics of inherited bone marrow failure and myelodysplasia Michael Yu Zhang Chair of the Supervisory Committee: Professor Mary-Claire King Department of Medicine (Medical Genetics) and Genome Sciences Bone marrow failure and myelodysplastic syndromes (BMF/MDS) are disorders of impaired blood cell production with increased leukemia risk. BMF/MDS may be acquired or inherited, a distinction critical for treatment selection. Currently, diagnosis of these inherited syndromes is based on clinical history, family history, and laboratory studies, which directs the ordering of genetic tests on a gene-by-gene basis. However, despite extensive clinical workup and serial genetic testing, many cases remain unexplained. We sought to define the genetic etiology and pathophysiology of unclassified bone marrow failure and myelodysplastic syndromes. First, to determine the extent to which patients remained undiagnosed due to atypical or cryptic presentations of known inherited BMF/MDS, we developed a massively-parallel, next- generation DNA sequencing assay to simultaneously screen for mutations in 85 BMF/MDS genes. Querying 71 pediatric and adult patients with unclassified BMF/MDS using this assay revealed 8 (11%) patients with constitutional, pathogenic mutations in GATA2 , RUNX1 , DKC1 , or LIG4 . All eight patients lacked classic features or laboratory findings for their syndromes. -

UCLA Electronic Theses and Dissertations
UCLA UCLA Electronic Theses and Dissertations Title Proteomic Analysis of Cancer Cell Metabolism Permalink https://escholarship.org/uc/item/8t36w919 Author Chai, Yang Publication Date 2013 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA Los Angeles Proteomic Analysis of Cancer Cell Metabolism A thesis submitted in partial satisfaction of the requirements of the degree Master of Science in Oral Biology by Yang Chai 2013 ABSTRACT OF THESIS Proteomic Analysis of Cancer Cell Metabolism by Yang Chai Master of Science in Oral Biology University of California, Los Angeles, 2013 Professor Shen Hu, Chair Tumor cells can adopt alternative metabolic pathways during oncogenesis. This is an event characterized by an enhanced utilization of glucose for rapid synthesis of macromolecules such as nucleotides, lipids and proteins. This phenomenon was also known as the ‘Warburg effect’, distinguished by a shift from oxidative phosphorylation to increased aerobic glycolysis in many types of cancer cells. Increased aerobic glycolysis was also indicated with enhanced lactate production and glutamine consumption, and has been suggested to confer growth advantage for proliferating cells during oncogenic transformation. Development of a tracer-based ii methodology to determine de novo protein synthesis by tracing metabolic pathways from nutrient utilization may certainly enhance current understanding of nutrient gene interaction in cancer cells. We hypothesized that the metabolic phenotype of cancer cells as characterized by nutrient utilization for protein synthesis is significantly altered during oncogenesis, and 13C stable isotope tracers may incorporate 13C into non-essential amino acids of protein peptides during de novo protein synthesis to reflect the underlying mechanisms in cancer cell metabolism. -

Mass Spectrometry Analysis Reveals Dynamic Changes of Protein
Japanese Journal of Gastroenterology and Hepatology Research Article Mass Spectrometry Analysis Reveals Dynamic Changes of Protein Components in Lipid Rafts from Rat Liver after Partial Hepatectomy Zhu B1*, Li J1*, Yang C1, Jiang M1, Zhou Q1, Wang Y1, 2* and Chen P1, 2* 1Department of Gastroenterology, The National & Local Joint Engineering Laboratory of Animal Peptide Drug Development, College of Life Sciences, Hunan Normal University, Changsha 410081, P.R. China 2Department of Gastroenterology, State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life Sciences, Hunan Normal University, Changsha, 410081, P.R. China Received: 08 Apr 2020 1. Abstract Accepted: 27 Apr 2020 Lipid raft, as scaffolding platform for signal transduction, plays important role in the liver re- Published: 30 Apr 2020 generation. But the lipid raft protein expression pattern during liver regeneration has been not * Corresponding author: reported. In this study, lipid raft proteins from the liver of 72 h post partial hepatectomy and Ping Chen, Department of Gastroen- sham-operated group were identified by liquid chromatography-tandem mass spectrometry (LC- terology, The National & Local Joint MS/MS) combined with label-free semi-quantitative analysis. Totally 458 lipid raft proteins were Engineering Laboratory of Animal identified, and most of identified lipid raft proteins were mainly involved in transporter, signal Peptide Drug Development, College transduction, and metabolism. Moreover, label-free quantification analysis suggested that the level of Life Sciences, Hunan Normal Uni- of 46 plasma membrane-related proteins have changed obviously (with ratio≥2) after 72 h hepa- versity, Changsha 410081, P.R. China, tectomy. Several differently expressed proteins, including caveolin-1 and flotillin-1, were validated E-mail: [email protected] by western blotting. -

Mapping Transmembrane Binding Partners for E-Cadherin Ectodomains
SUPPLEMENTARY INFORMATION TITLE: Mapping transmembrane binding partners for E-cadherin ectodomains. AUTHORS: Omer Shafraz 1, Bin Xie 2, Soichiro Yamada 1, Sanjeevi Sivasankar 1, 2, * AFFILIATION: 1 Department of Biomedical Engineering, 2 Biophysics Graduate Group, University of California, Davis, CA 95616. *CORRESPONDING AUTHOR: Sanjeevi Sivasankar, Tel: (530)-754-0840, Email: [email protected] Figure S1: Western blots a. EC-BioID, WT and Ecad-KO cell lysates stained for Ecad and tubulin. b. HRP-streptavidin staining of biotinylated proteins eluted from streptavidin coated magnetic beads incubated with cell lysates of EC-BioID with (+) and without (-) exogenous biotin. c. C-BioID, WT and Ecad-KO cell lysates stained for Ecad and tubulin. d. HRP-streptavidin staining of biotinylated proteins eluted from streptavidin coated magnetic beads incubated with cell lysates of C-BioID with (+) and without (-) exogenous biotin. (+) Biotin (-) Biotin Sample 1 Sample 2 Sample 3 Sample 4 Sample 1 Sample 2 Sample 3 Sample 4 Percent Percent Percent Percent Percent Percent Percent Percent Gene ID Coverage Coverage Coverage Coverage Coverage Coverage Coverage Coverage CDH1 29.6 31.4 41.1 36.5 10.8 6.7 28.8 29.1 DSG2 26 14.6 45 37 0.8 1.9 1.6 18.7 CXADR 30.2 26.2 32.7 27.1 0.0 0.0 0.0 6.9 EFNB1 24.3 30.6 24 30.3 0.0 0.0 0.0 0.0 ITGA2 16.5 22.2 30.1 33.4 1.1 1.1 5.2 7.2 CDH3 21.8 9.7 20.6 25.3 1.3 1.3 0.0 0.0 ITGB1 11.8 16.7 23.9 20.3 0.0 2.9 8.5 5.8 DSC3 9.7 7.5 11.5 13.3 0.0 0.0 2.6 0.0 EPHA2 23.2 31.6 31.6 30.5 0.8 0.0 0.0 5.7 ITGB4 21.8 27.8 33.1 30.7 0.0 1.2 3.9 4.4 ITGB3 23.5 22.2 26.8 24.7 0.0 0.0 5.2 9.1 CDH6 22.8 18.1 28.6 24.3 0.0 0.0 0.0 9.1 CDH17 8.8 12.4 20.7 18.4 0.0 0.0 0.0 0.0 ITGB6 12.7 10.4 14 17.1 0.0 0.0 0.0 1.7 EPHB4 11.4 8.1 14.2 16.3 0.0 0.0 0.0 0.0 ITGB8 5 10 15 17.6 0.0 0.0 0.0 0.0 ITGB5 6.2 9.5 15.2 13.8 0.0 0.0 0.0 0.0 EPHB2 8.5 4.8 9.8 12.1 0.0 0.0 0.0 0.0 CDH24 5.9 7.2 8.3 9 0.0 0.0 0.0 0.0 Table S1: EC-BioID transmembrane protein hits. -

Table S1 Changes in the Expression Levels of Genes Induced by Sevoflurane
Table S1 Changes in the expression levels of genes induced by sevoflurane * Gene ID Gene abbreviation log2 ratio Rtn4rl2 Reticulon 4 receptor-like 2 7.80 Mas1 MAS1 oncogene 7.76 Gpx6 Glutathione peroxidase 6 7.62 Ovol2 Ovo-like 2 (Drosophila) 7.19 Calcr Calcitonin receptor 6.81 Impg1 Interphotoreceptor matrix proteoglycan 1 6.72 Neurod6 Neurogenic differentiation 6 6.33 Neurod2 Neurogenic differentiation 2 6.30 Dio3 Deiodinase, iodothyronine type III 5.91 Cd6 CD6 antigen 5.80 Gucy2g Guanylate cyclase 2g 5.75 Jsrp1 Junctional sarcoplasmic reticulum protein 1 5.56 Slc38a4 Solute carrier family 38, member 4 5.51 Tmprss6 Transmembrane serine protease 6 5.47 Sh3rf2 SH3 domain containing ring finger 2 5.41 Ccbp2 Chemokine binding protein 2 5.33 Gprc5a G protein-coupled receptor, family C, group 5, member A 5.27 Clspn Claspin homolog (Xenopus laevis) 5.21 Robo3 Roundabout homolog 3 (Drosophila) 5.21 Figf C-fos induced growth factor 5.20 Trpc6 Transient receptor potential cation channel, subfamily C, member 6 5.19 Adora2a Adenosine A2a receptor 5.12 Tbx15 T-box 15 5.07 Cdsn Corneodesmosin 5.02 Cd4 CD4 antigen 5.02 Col19a1 Collagen, type XIX, alpha 1 4.98 Kcnh3 Potassium voltage-gated channel, subfamily H (eag-related), member 3 4.94 Gpr88 G-protein coupled receptor 88 4.90 C1qtnf7 C1q and tumor necrosis factor related protein 7 4.88 Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, Serpina9 antitrypsin), member 9 4.87 Il20ra Interleukin 20 receptor, alpha 4.79 Lrg1 Leucine-rich alpha-2-glycoprotein 1 4.78 Rpe65 Retinal -

Filaments and Phenotypes
F1000Research 2019, 8(F1000 Faculty Rev):1703 Last updated: 30 SEP 2019 REVIEW Filaments and phenotypes: cellular roles and orphan effects associated with mutations in cytoplasmic intermediate filament proteins [version 1; peer review: 2 approved] Michael W. Klymkowsky Molecular, Cellular & Developmental Biology, University of Colorado, Boulder, Boulder, CO, 80303, USA First published: 30 Sep 2019, 8(F1000 Faculty Rev):1703 ( Open Peer Review v1 https://doi.org/10.12688/f1000research.19950.1) Latest published: 30 Sep 2019, 8(F1000 Faculty Rev):1703 ( https://doi.org/10.12688/f1000research.19950.1) Reviewer Status Abstract Invited Reviewers Cytoplasmic intermediate filaments (IFs) surround the nucleus and are 1 2 often anchored at membrane sites to form effectively transcellular networks. Mutations in IF proteins (IFps) have revealed mechanical roles in epidermis, version 1 muscle, liver, and neurons. At the same time, there have been phenotypic published surprises, illustrated by the ability to generate viable and fertile mice null for 30 Sep 2019 a number of IFp-encoding genes, including vimentin. Yet in humans, the vimentin (VIM) gene displays a high probability of intolerance to loss-of-function mutations, indicating an essential role. A number of subtle F1000 Faculty Reviews are written by members of and not so subtle IF-associated phenotypes have been identified, often the prestigious F1000 Faculty. They are linked to mechanical or metabolic stresses, some of which have been found commissioned and are peer reviewed before to be ameliorated by the over-expression of molecular chaperones, publication to ensure that the final, published version suggesting that such phenotypes arise from what might be termed “orphan” effects as opposed to the absence of the IF network per se, an idea is comprehensive and accessible. -
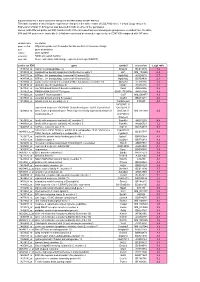
Supplementary File 1
Supplementary file 1: Gene expression changes in the white matter of CD47 KO mice This table contains a list of all gene expression changes in the white matter of CD47 KO mice ( > 2-fold: |Log2 ratio| > 1) Expression of total 14,875 genes was detected in both or either of the genotypes. Genes (with different probe set ID#) found in both of the increased and decreased gene groups were excluded from the table. 594 and 548 genes were markedly (> 2-fold) increased and decreased, respectively, in CD47 KO compared with WT mice. column name description probe set ID# Affymetrix probe set ID number for Mouse 430 2.0 Genome Arrays gene gene description symbol gene symbol accession NCBI accession number Log2 ratio Gene expression fold change expressed as Log2 (KO/WT) probe set ID# gene symbol accession Log2 ratio 1449153_at matrix metallopeptidase 12 Mmp12 BC019135 7.2 1419534_at oxidized low density lipoprotein (lectin-like) receptor 1 Olr1 NM_138648 6.2 1444176_at ATPase, H+ transporting, lysosomal V0 subunit D2 Atp6v0d2 AV204216 5.7 1434798_at ATPase, H+ transporting, lysosomal V0 subunit D2 Atp6v0d2 BB769890 2.3 1420504_at solute carrier family 6 (neurotransmitter transporter), member 14 Slc6a14 AF320226 5.5 1459934_at ubiquitin specific peptidase 42 Usp42 AA516835 5.2 1431800_at von Willebrand factor A domain containing 8 Vwa8 AK004956 5.2 1431927_at RIKEN cDNA 5033417F24 gene 5033417F24Rik AK018199 4.9 1419202_at cystatin F (leukocystatin) Cst7 NM_009977 4.9 1439943_at vacuolar protein sorting 54 (yeast) Vps54 BB201271 4.6 1419966_at tubulin,