High-Throughput Brain Activity Mapping and Machine Learning As a Foundation for Systems Neuropharmacology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
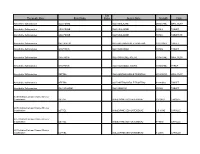
Therapeutic Class Brand Name P a Status Generic
P A Therapeutic Class Brand Name Status Generic Name Strength Form Absorbable Sulfonamides AZULFIDINE SULFASALAZINE 250MG/5ML ORAL SUSP Absorbable Sulfonamides AZULFIDINE SULFASALAZINE 500MG TABLET Absorbable Sulfonamides AZULFIDINE SULFASALAZINE 500MG TABLET DR Absorbable Sulfonamides BACTRIM DS SULFAMETHOXAZOLE/TRIMETHO 800-160MG TABLET Absorbable Sulfonamides GANTRISIN SULFISOXAZOLE 500MG TABLET Absorbable Sulfonamides GANTRISIN SULFISOXAZOLE ACETYL 500MG/5ML ORAL SUSP Absorbable Sulfonamides GANTRISIN SULFISOXAZOLE ACETYL 500MG/5ML SYRUP Absorbable Sulfonamides SEPTRA SULFAMETHOXAZOLE/TRIMETHO 200-40MG/5 ORAL SUSP Absorbable Sulfonamides SEPTRA SULFAMETHOXAZOLE/TRIMETHO 400-80MG TABLET Absorbable Sulfonamides SULFADIAZINE SULFADIAZINE 500MG TABLET ACE Inhibitor/Calcium Channel Blocker Combination LOTREL AMLODIPINE BESYLATE/BENAZ 10-20MG CAPSULE ACE Inhibitor/Calcium Channel Blocker Combination LOTREL AMLODIPINE BESYLATE/BENAZ 2.5-10MG CAPSULE ACE Inhibitor/Calcium Channel Blocker Combination LOTREL AMLODIPINE BESYLATE/BENAZ 5-10MG CAPSULE ACE Inhibitor/Calcium Channel Blocker Combination LOTREL AMLODIPINE BESYLATE/BENAZ 5-20MG CAPSULE P A Therapeutic Class Brand Name Status Generic Name Strength Form ACE Inhibitor/Calcium Channel Blocker Combination LOTREL AMLODIPINE BESYLATE/BENAZ 5-40MG CAPSULE ACE Inhibitor/Calcium Channel Blocker Combination LOTREL AMLODIPINE BESYLATE/BENAZ 10-40MG CAPSULE Acne Agents, Systemic ACCUTANE ISOTRETINOIN 10MG CAPSULE Acne Agents, Systemic ACCUTANE ISOTRETINOIN 20MG CAPSULE Acne Agents, Systemic ACCUTANE -

Classification Decisions Taken by the Harmonized System Committee from the 47Th to 60Th Sessions (2011
CLASSIFICATION DECISIONS TAKEN BY THE HARMONIZED SYSTEM COMMITTEE FROM THE 47TH TO 60TH SESSIONS (2011 - 2018) WORLD CUSTOMS ORGANIZATION Rue du Marché 30 B-1210 Brussels Belgium November 2011 Copyright © 2011 World Customs Organization. All rights reserved. Requests and inquiries concerning translation, reproduction and adaptation rights should be addressed to [email protected]. D/2011/0448/25 The following list contains the classification decisions (other than those subject to a reservation) taken by the Harmonized System Committee ( 47th Session – March 2011) on specific products, together with their related Harmonized System code numbers and, in certain cases, the classification rationale. Advice Parties seeking to import or export merchandise covered by a decision are advised to verify the implementation of the decision by the importing or exporting country, as the case may be. HS codes Classification No Product description Classification considered rationale 1. Preparation, in the form of a powder, consisting of 92 % sugar, 6 % 2106.90 GRIs 1 and 6 black currant powder, anticaking agent, citric acid and black currant flavouring, put up for retail sale in 32-gram sachets, intended to be consumed as a beverage after mixing with hot water. 2. Vanutide cridificar (INN List 100). 3002.20 3. Certain INN products. Chapters 28, 29 (See “INN List 101” at the end of this publication.) and 30 4. Certain INN products. Chapters 13, 29 (See “INN List 102” at the end of this publication.) and 30 5. Certain INN products. Chapters 28, 29, (See “INN List 103” at the end of this publication.) 30, 35 and 39 6. Re-classification of INN products. -

Design, Synthesis, and Evaluation of Antiepileptic Compounds Based on Β-Alanine and Isatin
Design, Synthesis, and Evaluation of Antiepileptic Compounds Based on β-Alanine and Isatin by Robert Philip Colaguori A thesis submitted in conformity with the requirements for the degree of Master of Science Department of Pharmaceutical Sciences University of Toronto © Copyright by Robert Philip Colaguori, 2016 ii Design, Synthesis, and Evaluation of Antiepileptic Compounds Based on β-Alanine and Isatin Robert Philip Colaguori Master of Science Department of Pharmaceutical Sciences University of Toronto 2016 Abstract Epilepsy is the fourth-most common neurological disorder in the world. Approximately 70% of cases can be controlled with therapeutics, however 30% remain pharmacoresistant. There is no cure for the disorder, and patients affected are subsequently medicated for life. Thus, there is a need to develop compounds that can treat not only the symptoms, but also delay/prevent progression. Previous work resulted in the discovery of NC-2505, a substituted β-alanine with activity against chemically induced seizures. Several N- and α-substituted derivatives of this compound were synthesized and evaluated in the kindling model and 4-AP model of epilepsy. In the kindling model, RC1-080 and RC1-102 were able to decrease the mean seizure score from 5 to 3 in aged mice. RC1-085 decreased the interevent interval by a factor of 2 in the 4-AP model. Future studies are focused on the synthesis of further compounds to gain insight on structure necessary for activity. iii Acknowledgments First and foremost, I would like to thank my supervisor Dr. Donald Weaver for allowing me to join the lab as a graduate student and perform the work ultimately resulting in this thesis. -

Stems for Nonproprietary Drug Names
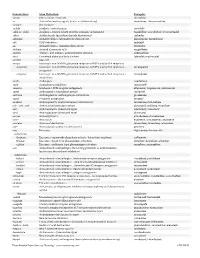
USAN STEM LIST STEM DEFINITION EXAMPLES -abine (see -arabine, -citabine) -ac anti-inflammatory agents (acetic acid derivatives) bromfenac dexpemedolac -acetam (see -racetam) -adol or analgesics (mixed opiate receptor agonists/ tazadolene -adol- antagonists) spiradolene levonantradol -adox antibacterials (quinoline dioxide derivatives) carbadox -afenone antiarrhythmics (propafenone derivatives) alprafenone diprafenonex -afil PDE5 inhibitors tadalafil -aj- antiarrhythmics (ajmaline derivatives) lorajmine -aldrate antacid aluminum salts magaldrate -algron alpha1 - and alpha2 - adrenoreceptor agonists dabuzalgron -alol combined alpha and beta blockers labetalol medroxalol -amidis antimyloidotics tafamidis -amivir (see -vir) -ampa ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) subgroup: -ampanel antagonists becampanel -ampator modulators forampator -anib angiogenesis inhibitors pegaptanib cediranib 1 subgroup: -siranib siRNA bevasiranib -andr- androgens nandrolone -anserin serotonin 5-HT2 receptor antagonists altanserin tropanserin adatanserin -antel anthelmintics (undefined group) carbantel subgroup: -quantel 2-deoxoparaherquamide A derivatives derquantel -antrone antineoplastics; anthraquinone derivatives pixantrone -apsel P-selectin antagonists torapsel -arabine antineoplastics (arabinofuranosyl derivatives) fazarabine fludarabine aril-, -aril, -aril- antiviral (arildone derivatives) pleconaril arildone fosarilate -arit antirheumatics (lobenzarit type) lobenzarit clobuzarit -arol anticoagulants (dicumarol type) dicumarol -

Modifications to the Harmonized Tariff Schedule of the United States To
U.S. International Trade Commission COMMISSIONERS Shara L. Aranoff, Chairman Daniel R. Pearson, Vice Chairman Deanna Tanner Okun Charlotte R. Lane Irving A. Williamson Dean A. Pinkert Address all communications to Secretary to the Commission United States International Trade Commission Washington, DC 20436 U.S. International Trade Commission Washington, DC 20436 www.usitc.gov Modifications to the Harmonized Tariff Schedule of the United States to Implement the Dominican Republic- Central America-United States Free Trade Agreement With Respect to Costa Rica Publication 4038 December 2008 (This page is intentionally blank) Pursuant to the letter of request from the United States Trade Representative of December 18, 2008, set forth in the Appendix hereto, and pursuant to section 1207(a) of the Omnibus Trade and Competitiveness Act, the Commission is publishing the following modifications to the Harmonized Tariff Schedule of the United States (HTS) to implement the Dominican Republic- Central America-United States Free Trade Agreement, as approved in the Dominican Republic-Central America- United States Free Trade Agreement Implementation Act, with respect to Costa Rica. (This page is intentionally blank) Annex I Effective with respect to goods that are entered, or withdrawn from warehouse for consumption, on or after January 1, 2009, the Harmonized Tariff Schedule of the United States (HTS) is modified as provided herein, with bracketed matter included to assist in the understanding of proclaimed modifications. The following supersedes matter now in the HTS. (1). General note 4 is modified as follows: (a). by deleting from subdivision (a) the following country from the enumeration of independent beneficiary developing countries: Costa Rica (b). -

Pdf4 Complex I and the Aryl Palladium Precursor II Underwent Sequential Single Electron Abstraction from Aryl Pd(II) Complex
Design, synthesis, methodology development, and evaluation of PET imaging agents targeting cancer and CNS disorders By Gengyang Yuan B.S. in Chemical Engineering and Technology, Zhejiang University of Technology M.S. in Pharmaceutical Engineering, Zhejiang University A dissertation submitted to The Faculty of the College of Science of Northeastern University in partial fulfillment of the requirements for the degree of Doctor of Philosophy April 21, 2017 Dissertation directed by Michael P. Pollastri Associate Professor and Chair of Chemistry and Chemical Biology Co-directed by Neil Vasdev Adjunct Associate Professor of Chemsitry and Chemical Biology Associate Professor of Radiology, Massachusetts General Hospital and Harvard Medical School Dedication To my parents Zhijun and Yongmian and my wife Ran and daughter Isabella ii Acknowledgements This dissertation would not have been possible without the support, guidance and encouragement of numerous people who have helped me along the way. First and foremost, I would like to thank Northeastern University and the Department of Chemistry and Chemical Biology for supporting me to pursue my doctoral study. I would like to especially thank my current advisor Professor Michael Pollastri for helping me out when I needed it the most. I appreciate you for taking me into your group and giving me full support to finish my thesis projects. I also especially thank my co-advisor Professor Neil Vasdev for taking me into his group at Mass. General Hospital & Harvard Medical School and teaching me the PET radiochemistry and PET imaging. I could not image how I could accomplish this work without your help. I also got a lot of help from Dr. -

"2-Oxo-1-Pyrrolidine Derivative and Its Pharmaceutical Uses"
(19) & (11) EP 1 452 524 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C07D 207/27 (2006.01) A61K 31/4015 (2006.01) 14.10.2009 Bulletin 2009/42 A61P 25/08 (2006.01) (21) Application number: 04007878.4 (22) Date of filing: 21.02.2001 (54) "2-oxo-1-pyrrolidine derivative and its pharmaceutical uses" "2-Oxo-1-Pyrrolidinderivate und ihre pharmazeutische Verwendung" "Dérivé de 2-oxo-1-pyrrolidine et ses applications pharmaceutique" (84) Designated Contracting States: • Talaga, Patrice AT BE CH CY DE DK ES FI FR GB GR IE IT LI LU 1170 Watermael-Boitsfort (BE) MC NL PT SE TR (74) Representative: UCB (30) Priority: 23.02.2000 GB 0004297 Intellectual Property c/o UCB S.A. (43) Date of publication of application: Intellectual Property Department 01.09.2004 Bulletin 2004/36 Allée de la Recherche 60 1070 Brussels (BE) (62) Document number(s) of the earlier application(s) in accordance with Art. 76 EPC: (56) References cited: 01925354.1 / 1 265 862 WO-A-99/13911 GB-A- 1 309 692 (73) Proprietor: UCB Pharma, S.A. • PROUS: "LEVETIRACETAM" DRUGS OF THE 1170 Brussels (BE) FUTURE, BARCELONA, ES, vol. 19, no. 2, 1994, pages 111-113, XP000908882 ISSN: 0377-8282 (72) Inventors: • FISCHER W ET AL: "Die Wirksamkeit von • Differding, Edmond Piracetam, Meclofenoxat und Vinpocetin in 1348 Ottignies-Louvain-La-Neuve (BE) verschiedenen Krampfmodellen bei der Maus" • Kenda, Benoît PHARMAZIE, VEB VERLAG VOLK UND 5080 Emines (BE) GESUNDHEIT. BERLIN, DD, vol. 46, no. -
Downloaded from Survive Nursing | Survivenursing.Com V20110426
Generic Stem Stem Definition Examples -abine (see -arabine, -citabine) decitabine -ac Anti-inflammatory agents (acetic acid derivatives) bromfenac; dexpemedolac -acetam See -racetam -actide Synthetic corticotropins seractide -adol or -aldol- Analgesics (mixed opiate receptor agonists/ antagonists) tazadolene; spiradolene; levonantradol -adox Antibacterials (quinoline dioxide derivatives) carbadox -afenone Antiarrhythmics (propafenone derivatives) alprafenone; diprafenone -afil PDE5 inhibitors tadalafil -aj- Antiarrhythmics (ajmaline derivatives) lorajmine -aldrate Antacid aluminum salts magaldrate -algron Alpha1 - and alpha2 - adrenoreceptor agonists dabuzalgron -alol Combined alpha and beta blockers labetalol; medroxalol -amivir (see -vir) -ampa Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) -ampanel Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) ; becampanel antagonists -ampator Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) ; forampator modulators -andr- Androgens nandrolone -anib Angiogenesis inhibitors semaxanib -anserin Serotonin 5-HT2 receptor antagonists altanserin; tropanserin; adatanserin -antel Anthelmintics (undefined group) carbantel -antrone Antineoplastics; anthraquinone derivatives pixantrone -apsel P-selectin antagonists torapsel -arabine Antineoplastics (arabinofuranosyl derivatives) fazarabine; fludarabine aril-, -aril, -aril- Antiviral (arildone derivatives) pleconaril; arildone; fosarilate -arit Antirheumatics (lobenzarit type) lobenzarit; clobuzarit -arol -

(12) Patent Application Publication (10) Pub. No.: US 2015/0202317 A1 Rau Et Al
US 20150202317A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0202317 A1 Rau et al. (43) Pub. Date: Jul. 23, 2015 (54) DIPEPTDE-BASED PRODRUG LINKERS Publication Classification FOR ALPHATIC AMNE-CONTAINING DRUGS (51) Int. Cl. A647/48 (2006.01) (71) Applicant: Ascendis Pharma A/S, Hellerup (DK) A638/26 (2006.01) A6M5/9 (2006.01) (72) Inventors: Harald Rau, Heidelberg (DE); Torben A 6LX3/553 (2006.01) Le?mann, Neustadt an der Weinstrasse (52) U.S. Cl. (DE) CPC ......... A61K 47/48338 (2013.01); A61 K3I/553 (2013.01); A61 K38/26 (2013.01); A61 K (21) Appl. No.: 14/674,928 47/48215 (2013.01); A61M 5/19 (2013.01) (22) Filed: Mar. 31, 2015 (57) ABSTRACT The present invention relates to a prodrug or a pharmaceuti Related U.S. Application Data cally acceptable salt thereof, comprising a drug linker conju (63) Continuation of application No. 13/574,092, filed on gate D-L, wherein D being a biologically active moiety con Oct. 15, 2012, filed as application No. PCT/EP2011/ taining an aliphatic amine group is conjugated to one or more 050821 on Jan. 21, 2011. polymeric carriers via dipeptide-containing linkers L. Such carrier-linked prodrugs achieve drug releases with therapeu (30) Foreign Application Priority Data tically useful half-lives. The invention also relates to pharma ceutical compositions comprising said prodrugs and their use Jan. 22, 2010 (EP) ................................ 10 151564.1 as medicaments. US 2015/0202317 A1 Jul. 23, 2015 DIPEPTDE-BASED PRODRUG LINKERS 0007 Alternatively, the drugs may be conjugated to a car FOR ALPHATIC AMNE-CONTAINING rier through permanent covalent bonds. -

DILANTIN Oral Suspension Page 1 of 23 LPDPYC102019 Pfleet Number: 2019-0051655, 2019-0053053 4.2 Posology and Method of Administration
For the use only of a Registered Medical Practitioners or a Hospital or a Laboratory Phenytoin Oral Suspension USP DILANTIN Suspension 1. NAME OF MEDICINAL PRODUCT DILANTIN SUSPENSION 2. QUALITATIVE AND QUANTITATIVE COMPOSITION Phenytoin is an anticonvulsant drug, related to the barbiturates in chemical structure, but has a five-membered ring. The chemical name is 5,5-diphenyl-2,4-imidazolidinedione. Each 5 mL of oral suspension contains 125 mg of phenytoin I.P. All strengths mentioned in this document might not be available in the market. For full list of excipients please see section 6.1. 3. PHARMACEUTICAL FORM Oral suspension 4. CLINICAL PARTICULARS 4.1 Therapeutic Indications Phenytoin is indicated for the control of generalized tonic-clonic (grand mal) and complex partial (psychomotor, temporal lobe) seizures and prevention and treatment of seizures occurring during or following neurosurgery. Trademark of Parke, Davis & Company, LLC, USA. Licensed user: Pfizer Limited, India DILANTIN Oral Suspension Page 1 of 23 LPDPYC102019 PfLEET Number: 2019-0051655, 2019-0053053 4.2 Posology and Method of Administration General NOT FOR PARENTERAL USE Phenytoin capsules and solution for injection are formulated with the sodium salt of phenytoin. The free acid form of phenytoin is used in the phenytoin suspensions (30 mg/5 mL (pediatric) and 125 mg/5 mL). Because there is approximately an 8% increase in drug content with the free acid form over that of the sodium salt, dosage adjustments and serum level monitoring may be necessary when switching from a product formulated with the free acid to a product formulated with the sodium salt and vice versa. -

Reproductive Performance and Embriotoxicity of Rats Exposed to Carbamazepine
Revista Brasileira de Ciências Farmacêuticas Brazilian Journal of Pharmaceutical Sciences vol. 44, n. 3, jul./set., 2008 Reproductive performance and embriotoxicity of rats exposed to carbamazepine Marli Gerenutti*, Carolina Clavijos de Oliveira, Aliana Conceição Ribeiro de Miranda, Raquel Mendonça Rosa, Fernando de Sá Del Fiol Curso de Farmácia, Universidade de Sorocaba Uniterms To study the possible effects of carbamazepine in rats during • Carbamazepine pregnancy and fetuses´ physical development, carbamazepine • Teratology solubilized in propilenglycol was orally administered (20 and • Embriotoxicity 40 mg/kg) to the female rats from the 2nd to the 19th days of • Pregnancy pregnancy. Propilenglycol was administered to the control group. • Reproduction The animals were sacrificed on the 20th day, when 50% of the offspring were fixed in Bouin´s solution and the remaining 50% were submitted to diaphanization. The carbamazepine administration caused reduction on weight gain of pregnant rats and did not damage the females´ reproductive performance. In the fetuses´ physical development, it was observed a flattening on the skull soft tissues and bones; delay in the skull bone development; cartilage calcification increase between hip and *Correspondência: femur and reduction in the number of the sternum ossifications. M.Gerenutti, Universidade de Sorocaba Although carbamazepine has not caused general changes over Av. Dr. Eugênio Salerno, 140 female rats´ reproductive performance, it produced significant 18035-430 - Sorocaba - SP, Brasil E-mail [email protected]. alterations in the development of the fetuses´ skeletal parameters. INTRODUCTION to anatomical and/or biochemical abnormalities specially when specifying a group of epileptic pregnant females Drugs usage during pregnancy and breast feeding (Kaneko et al., 2001; Samren et al., 2003). -

Antiepileptic Drug Treatment in 1959
Antiepileptic Drug Treatment in 1959 The two decades between 1938 and 1958 were notable for a large number of new medicinal com- pounds.Phenytoin was of course the most important, but other drugs, some of which are still prescribed, were introduced at this time. Not only drugs – for the ketogenic diet was also popu- larised in this period. Lennox in his book in 1960 gave a long treatise on drug therapy. Head- ing his list were the bromides, which he found rather ineffective, but barbiturates were greeted enthusiastically. Lennox mentioned that 2,500 compounds had been synthesised, and of these 50 compounds were marketed of which phenobarbital was the most frequently used for epilepsy. He was also enthusiastic about phenytoin, especially for patients ‘with long–standing convul- sions previously unrelieved by phenobarbital’ although Mesantoin outranked it on several points, and indeed he favoured their combined use: ‘Mesantoin and Dilantin are Damon and Pythias in respect to their suitability for joint action. Similarity of action gives a doubled therapeutic effect; the dissimilarity of their side reactions keeps these within bounds.’ Phenacemide was ‘what in athletics might be called a triple threat because, more than any other drug, it acts against each of the three main types of seizures, and especially against the most feared psychomotor seizures. However, it is also a triple threat to the patient himself because of possible effect on the marrow, the liver or the psyche.’ Given one chance in 250 of not surviving this treatment, he Drugs introduced into clinical asked, ‘Is the risk too great?’ Trimethadione practice between 1938 and 1958 (Tridione) ‘heads the list of drugs that are 1938 Dilantin Phenytoin peculiarly beneficial to persons subject to 1941 Diamox Acetazolamide 1946 Tridione Trimethadione petits’.