Rrna of Alphaproteobacteria Rickettsiales and Mtdna Pattern Analyzing by Decision Tree
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Batch Learning Self-Organizing Maps), to the Microbiome Analysis of Ticks
Title A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks Nakao, Ryo; Abe, Takashi; Nijhof, Ard M; Yamamoto, Seigo; Jongejan, Frans; Ikemura, Toshimichi; Sugimoto, Author(s) Chihiro The ISME Journal, 7(5), 1003-1015 Citation https://doi.org/10.1038/ismej.2012.171 Issue Date 2013-03 Doc URL http://hdl.handle.net/2115/53167 Type article (author version) File Information ISME_Nakao.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks Ryo Nakao1,a, Takashi Abe2,3,a, Ard M. Nijhof4, Seigo Yamamoto5, Frans Jongejan6,7, Toshimichi Ikemura2, Chihiro Sugimoto1 1Division of Collaboration and Education, Research Center for Zoonosis Control, Hokkaido University, Kita-20, Nishi-10, Kita-ku, Sapporo, Hokkaido 001-0020, Japan 2Nagahama Institute of Bio-Science and Technology, Nagahama, Shiga 526-0829, Japan 3Graduate School of Science & Technology, Niigata University, 8050, Igarashi 2-no-cho, Nishi- ku, Niigata 950-2181, Japan 4Institute for Parasitology and Tropical Veterinary Medicine, Freie Universität Berlin, Königsweg 67, 14163 Berlin, Germany 5Miyazaki Prefectural Institute for Public Health and Environment, 2-3-2 Gakuen Kibanadai Nishi, Miyazaki 889-2155, Japan 6Utrecht Centre for Tick-borne Diseases (UCTD), Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands 7Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria, Private Bag X04, 0110 Onderstepoort, South Africa aThese authors contributed equally to this work. Keywords: BLSOMs/emerging diseases/metagenomics/microbiomes/symbionts/ticks Running title: Tick microbiomes revealed by BLSOMs Subject category: Microbe-microbe and microbe-host interactions Abstract Ticks transmit a variety of viral, bacterial and protozoal pathogens, which are often zoonotic. -

Babela Massiliensis, a Representative of a Widespread Bacterial
Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, Samia Benamar, Didier Raoult, Eugene V. Koonin, Bernard La Scola To cite this version: Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, et al.. Babela mas- siliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae. Biology Direct, BioMed Central, 2015, 10 (13), 10.1186/s13062-015-0043-z. hal-01217089 HAL Id: hal-01217089 https://hal-amu.archives-ouvertes.fr/hal-01217089 Submitted on 19 Oct 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Pagnier et al. Biology Direct (2015) 10:13 DOI 10.1186/s13062-015-0043-z RESEARCH Open Access Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier1, Natalya Yutin2, Olivier Croce1, Kira S Makarova2, Yuri I Wolf2, Samia Benamar1, Didier Raoult1, Eugene V Koonin2 and Bernard La Scola1* Abstract Background: Only a small fraction of bacteria and archaea that are identifiable by metagenomics can be grown on standard media. -

Characterization of the Interaction Between R. Conorii and Human
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 4-5-2018 Characterization of the Interaction Between R. Conorii and Human Host Vitronectin in Rickettsial Pathogenesis Abigail Inez Fish Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Bacteria Commons, Bacteriology Commons, Biology Commons, Immunology of Infectious Disease Commons, and the Pathogenic Microbiology Commons Recommended Citation Fish, Abigail Inez, "Characterization of the Interaction Between R. Conorii and Human Host Vitronectin in Rickettsial Pathogenesis" (2018). LSU Doctoral Dissertations. 4566. https://digitalcommons.lsu.edu/gradschool_dissertations/4566 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. CHARACTERIZATION OF THE INTERACTION BETWEEN R. CONORII AND HUMAN HOST VITRONECTIN IN RICKETTSIAL PATHOGENESIS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Interdepartmental Program in Biomedical and Veterinary Medical Sciences Through the Department of Pathobiological Sciences by Abigail Inez -

Transition of Serum Cytokine Concentration in Rickettsia Japonica Infection
Article Transition of Serum Cytokine Concentration in Rickettsia japonica Infection Makoto Kondo, Yoshiaki Matsushima, Kento Mizutani, Shohei Iida, Koji Habe and Keiichi Yamanaka * Department of Dermatology, Mie University Graduate School of medicine, 2-174 Edobashi, Tsu, Mie 514-8507, Japan; [email protected] (M.K.); [email protected] (Y.M.); [email protected] (K.M.); [email protected] (S.I.); [email protected] (K.H.) * Correspondence: [email protected]; Tel.: +81-59-231-5025; Fax: +81-59-231-5206 Received: 31 October 2020; Accepted: 8 December 2020; Published: 11 December 2020 Abstract: (1) Background. Rickettsia japonica (R. japonica) infection induces severe inflammation, and the disappearance of eosinophil in the acute stage is one of the phenomena. (2) Materials and Methods. In the current study, we measured the serum concentrations of Th1, Th2, and Th17 cytokines in the acute and recovery stages. (3) Results. In the acute phase, IL-6 and IFN-γ levels were elevated and we speculated that they played a role as a defense mechanism against R. japonica. The high concentration of IFN-γ suppressed the differentiation of eosinophil and induced apoptosis of eosinophil, leading to the disappearance of eosinophil. On day 7, IL-6 and IFN-γ concentrations were decreased, and Th2 cytokines such as IL-5 and IL-9 were slightly increased. On day 14, eosinophil count recovered to the normal level. The transition of serum cytokine concentration in R. japonica infection was presented. (4) Conclusions. -

Rickettsia Prowazekii Common Human Exposure Routes
APPENDIX 2 Rickettsia prowazekii Common Human Exposure Routes: Disease Agent: • Exposure to the feces of infected body lice. The lice are infected by a human blood meal. The rickettsiae • Rickettsia prowazekii reproduce in the louse gut epithelium. Infection occurs when louse feces are scratched into the skin, Disease Agent Characteristics: inoculated onto mucous membrane or inhaled. • As a bioweapon, the agent can be aerosolized, with • Rickettsiae are obligate intracellular Gram-negative intent of infection through inhalation. bacteria. • Sporadic cases occur after exposure to flying squir- • Order: Rickettsiales; Family: Rickettsiaceae rels, most likely as a result of exposure to squirrel flea • Size: 0.3 ¥ 1.0 mm intracellular bacteria that take up feces. The organism has also been identified in ticks Gram stain poorly feeding on livestock in Africa. • Nucleic acid: Rickettsial genomes are among the Likelihood of Secondary Transmission: smallest of bacteria at 1000-1600 kb. The R. prowazekii genome is 1100 kb. • No evidence of direct person-to-person transmission • Physicochemical properties: Susceptible to 1% • Under crowded conditions, where bathing and sodium hypochlorite, 70% ethanol, glutaraldehyde, washing clothes are difficult, and where lice are formaldehyde and quaternary ammonium disinfec- present, typhus can spread explosively. tants. Sensitive to moist heat (121°C for at least • Recent outbreaks have occurred in a number of areas 15 min) and dry heat (160-170°C for at least 1 hour). in the world under conditions of war and population The organism is stable in tick tissues or blood under displacement. ambient environmental conditions, surviving up to 1 year; sensitive to drying (feces of infected ticks At-Risk Populations: quickly lose their infectivity on drying). -

Sanguineus Bacterial Communities Rhipicephalus
Composition and Seasonal Variation of Rhipicephalus turanicus and Rhipicephalus sanguineus Bacterial Communities Itai Lalzar, Shimon Harrus, Kosta Y. Mumcuoglu and Yuval Gottlieb Appl. Environ. Microbiol. 2012, 78(12):4110. DOI: 10.1128/AEM.00323-12. Published Ahead of Print 30 March 2012. Downloaded from Updated information and services can be found at: http://aem.asm.org/content/78/12/4110 These include: SUPPLEMENTAL MATERIAL Supplemental material http://aem.asm.org/ REFERENCES This article cites 44 articles, 17 of which can be accessed free at: http://aem.asm.org/content/78/12/4110#ref-list-1 CONTENT ALERTS Receive: RSS Feeds, eTOCs, free email alerts (when new articles cite this article), more» on June 10, 2013 by guest Information about commercial reprint orders: http://journals.asm.org/site/misc/reprints.xhtml To subscribe to to another ASM Journal go to: http://journals.asm.org/site/subscriptions/ Composition and Seasonal Variation of Rhipicephalus turanicus and Rhipicephalus sanguineus Bacterial Communities Itai Lalzar,a Shimon Harrus,a Kosta Y. Mumcuoglu,b and Yuval Gottlieba Koret School of Veterinary Medicine, The Robert H. Smith Faculty of Agriculture, Food and Environment, The Hebrew University of Jerusalem, Rehovot, Israel,a and Department of Microbiology and Molecular Genetics, The Kuvin Center for the Study of Infectious and Tropical Diseases, Hadassah Medical School, The Institute for Medical Research Israel-Canada, The Hebrew University of Jerusalem, Jerusalem, Israelb A 16S rRNA gene approach, including 454 pyrosequencing and quantitative PCR (qPCR), was used to describe the bacterial com- munity in Rhipicephalus turanicus and to evaluate the dynamics of key bacterial tenants of adult ticks during the active questing season. -
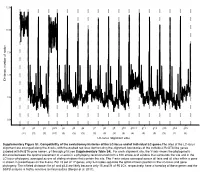
LC-Locus Alignment Sites Distance, Number of Nodes Supplementary
12.5 10.0 7.5 5.0 Distance, number of nodes 2.5 0.0 g1 g2 g3 g3.5 g4 g5 g6 g7 g8 g9 g10 g10.1 g11 g12 g13 g14 g15 (11) (3) (3) (10) (3) (3) (3) (3) (3) (3) (3) (4) (3) (3) (3) (2) (3) LC-locus alignment sites Supplementary Figure S1. Compatibility of the evolutionary histories of the LC-locus and of individual LC genes.The sites of the LC-locus alignment are arranged along the X-axis, with the dashed red lines demarcating the alignment boundaries of the individual RcGTA-like genes (labeled with RcGTA gene names, g1 through g15; see Supplementary Table S4). For each alignment site, the Y-axis shows the phylogenetic distance between the optimal placement of a taxon in a phylogeny reconstructed from a 100 amino-acid window that surrounds the site and in the LC-locus phylogeny, averaged across all sliding windows that contain the site. The Y-axis values averaged across all taxa and all sites within a gene is shown in parentheses on the X-axis. For 15 out of 17 genes, only 2-4 nodes separate the optimal taxon position in the LC-locus and gene phylogeny. The inflated distances for g1 and g3.5 are likely because only 15 and 21 of 95 LCs, respectively, have a homolog of these genes and the SSPB analysis is highly sensitive to missing data (Berger et al. 2011). a. Bacteria Unassigned Thermotogae Tenericutes Synergistetes Spirochaetes Proteobacteria ylum Planctomycetes h p Firmicutes Deferribacteres Cyanobacteria Chloroflexi Bacteroidetes Actinobacteria Acidobacteria 1(11,750) 2(1,750) 3(2,538) 4(168) 5(51) 6(54) 7(43) 8(32) 9(26) 10(40) 11(33) 12(198) 13(173) 14(101) 15(98) 16(43) 17(114) Number of rcc01682−rcc01698 homologs in a cluster b. -

Genome Project Reveals a Putative Rickettsial Endosymbiont
GBE Bacterial DNA Sifted from the Trichoplax adhaerens (Animalia: Placozoa) Genome Project Reveals a Putative Rickettsial Endosymbiont Timothy Driscoll1,y, Joseph J. Gillespie1,2,*,y, Eric K. Nordberg1,AbduF.Azad2, and Bruno W. Sobral1,3 1Virginia Bioinformatics Institute at Virginia Polytechnic Institute and State University 2Department of Microbiology and Immunology, University of Maryland School of Medicine 3Present address: Nestle´ Institute of Health Sciences SA, Campus EPFL, Quartier de L’innovation, Lausanne, Switzerland *Corresponding author: E-mail: [email protected]. yThese authors contributed equally to this work. Accepted: March 1, 2013 Abstract Eukaryotic genome sequencing projects often yield bacterial DNA sequences, data typically considered as microbial contamination. However, these sequences may also indicate either symbiont genes or lateral gene transfer (LGT) to host genomes. These bacterial sequences can provide clues about eukaryote–microbe interactions. Here, we used the genome of the primitive animal Trichoplax adhaerens (Metazoa: Placozoa), which is known to harbor an uncharacterized Gram-negative endosymbiont, to search for the presence of bacterial DNA sequences. Bioinformatic and phylogenomic analyses of extracted data from the genome assembly (181 bacterial coding sequences [CDS]) and trace read archive (16S rDNA) revealed a dominant proteobacterial profile strongly skewed to Rickettsiales (Alphaproteobacteria) genomes. By way of phylogenetic analysis of 16S rDNA and 113 proteins conserved across proteobacterial genomes, as well as identification of 27 rickettsial signature genes, we propose a Rickettsiales endosymbiont of T. adhaerens (RETA). The majority (93%) of the identified bacterial CDS belongs to small scaffolds containing prokaryotic-like genes; however, 12 CDS were identified on large scaffolds comprised of eukaryotic-like genes, suggesting that T. -

Rickettsialpox-A Newly Recognized Rickettsial Disease V
Public Health Reports Vol. 62 * MAY 30, 1947 * No. 22 Printed With the Approval of the Bureau of the Budget as Required by Rule 42 of the Joint-Committee on Printing RICKETTSIALPOX-A NEWLY RECOGNIZED RICKETTSIAL DISEASE V. RECOVERY OF RICKETTSIA AKARI FROM A HOUSE MOUSE (MUS MUSCULUS)1 By ROBERT J. HUEBNER, Senior Assistant Surgeon, WILLIAm L. JELLISON, Parasitologist, CHARLES ARMSTRONG, Medical Director, United States Public Health Service Ricketttia akari, the causative agent of rickettsialpox, was isolated from the blood of persons ill with this disease (1) and from rodent mites Allodermanyssus sanguineus Hirst inhabiting the domicile of ill per- sons (2). This paper describes the isolation of R. akari from a house mouse (Mus musculus) trapped on the same premises-a housing development in the citr of New York where more than 100 cases of rickettsialpox have occurred (3), (4), (5), (6). Approximately 60 house mice were trapped in the basements of this housing development where rodent harborage existed in store rooms and in incinerator ashpits. Engorged mites were occasionally found attached to the mice, the usual site of attachment being the rump. Mites were frequently found inside the box traps after the captured mice were removed. Early attempts to isolate the etiological agent of rickettisalpox from these mice were complicated by the presence of choriomeningitis among them. Twelve successive suspensions of mouse tissue, repre- senting 16 house mice, inoculated intracerebrally into laboratory mice (Swiss strain) and intraperitoneally into guinea pigs resulted in the production of a highly lethal disease in both species which was identified immunologically as choriomeningitis. -

Ohio Department of Health, Bureau of Infectious Diseases Disease Name Class A, Requires Immediate Phone Call to Local Health
Ohio Department of Health, Bureau of Infectious Diseases Reporting specifics for select diseases reportable by ELR Class A, requires immediate phone Susceptibilities specimen type Reportable test name (can change if Disease Name other specifics+ call to local health required* specifics~ state/federal case definition or department reporting requirements change) Culture independent diagnostic tests' (CIDT), like BioFire panel or BD MAX, E. histolytica Stain specimen = stool, bile results should be sent as E. histolytica DNA fluid, duodenal fluid, 260373001^DETECTED^SCT with E. histolytica Antigen Amebiasis (Entamoeba histolytica) No No tissue large intestine, disease/organism-specific DNA LOINC E. histolytica Antibody tissue small intestine codes OR a generic CIDT-LOINC code E. histolytica IgM with organism-specific DNA SNOMED E. histolytica IgG codes E. histolytica Total Antibody Ova and Parasite Anthrax Antibody Anthrax Antigen Anthrax EITB Acute Anthrax EITB Convalescent Anthrax Yes No Culture ELISA PCR Stain/microscopy Stain/spore ID Eastern Equine Encephalitis virus Antibody Eastern Equine Encephalitis virus IgG Antibody Eastern Equine Encephalitis virus IgM Arboviral neuroinvasive and non- Eastern Equine Encephalitis virus RNA neuroinvasive disease: Eastern equine California serogroup virus Antibody encephalitis virus disease; LaCrosse Equivocal results are accepted for all California serogroup virus IgG Antibody virus disease (other California arborviral diseases; California serogroup virus IgM Antibody specimen = blood, serum, serogroup -

Rickettsia Helvetica in Dermacentor Reticulatus Ticks
DISPATCHES The Study Rickettsia helvetica Using the cloth-dragging method, during March–May 2007 we collected 100 adult Dermacentor spp. ticks from in Dermacentor meadows in 2 different locations near Cakovec, between the Drava and Mura rivers in the central part of Medjimurje Coun- reticulatus Ticks ty. This area is situated in the northwestern part of Croatia, at Marinko Dobec, Dragutin Golubic, 46″38′N, 16″43′E, and has a continental climate with an Volga Punda-Polic, Franz Kaeppeli, average annual air temperature of 10.4°C at an altitude of and Martin Sievers 164 m. To isolate DNA from ticks, we modifi ed the method We report on the molecular evidence that Dermacentor used by Nilsson et al. (11). Before DNA isolation, ticks reticulatus ticks in Croatia are infected with Rickettsia hel- were disinfected in 70% ethanol and dried. Each tick was vetica (10%) or Rickettsia slovaca (2%) or co-infected with mechanically crushed in a Dispomix 25 tube with lysis buf- both species (1%). These fi ndings expand the knowledge of fer by using the Dispomix (Medic Tools, Zug, Switzerland). the geographic distribution of R. helvetica and D. reticulatus Lysis of each of the crushed tick samples was carried out in ticks. a solution of 6.7% sucrose, 0.2% proteinase K, 20 mg/mL lysozyme, and 10 ng/ml RNase A for 16 h at 37°C; 0.5 mo- ickettsia helvetica organisms were fi rst isolated from lar EDTA, and 20% sodium dodecyl sulfate was added and RIxodes ricinus ticks in Switzerland and were consid- further incubated for 1 h at 37°C. -

BEI Resources Product Information Sheet Catalog No. NR-51407 Rickettsia Helvetica, Strain C3
Product Information Sheet for NR-51407 Rickettsia helvetica, Strain C3 Citation: Acknowledgment for publications should read “The following Catalog No. NR-51407 reagent was obtained through BEI Resources, NIAID, NIH: Rickettsia helvetica, Strain C3, NR-51407.” For research use only. Not for human use. Biosafety Level: 3 Contributor: Appropriate safety procedures should always be used with this ATCC® material. Laboratory safety is discussed in the following publication: U.S. Department of Health and Human Services, Manufacturer: Public Health Service, Centers for Disease Control and BEI Resources Prevention, and National Institutes of Health. Biosafety in Microbiological and Biomedical Laboratories. 5th ed. Product Description: Washington, DC: U.S. Government Printing Office, 2009; see Bacteria Classification: Rickettsiaceae, Rickettsia www.cdc.gov/biosafety/publications/bmbl5/index.htm. Species: Rickettsia helvetica (also known as Swiss agent)1,2 Strain: C3 Disclaimers: Original Source: Rickettsia helvetica (R. helvetica), strain C3 You are authorized to use this product for research use only. was isolated from triturated Ixodes ricinus (I. ricinus) It is not intended for human use. nymphs from Switzerland in 1979.1,2 Use of this product is subject to the terms and conditions of R. helvetica is a member of the spotted fever group of the BEI Resources Material Transfer Agreement (MTA). The Rickettsiae found in Europe and Asia.3,4 R. helvetica is an MTA is available on our Web site at www.beiresources.org. intracellular Gram-negative pathogen that is transmitted to a human host via interaction with an infected tick (commonly While BEI Resources uses reasonable efforts to include I. ricinus but has also been isolated from Dermacentor accurate and up-to-date information on this product sheet, ® reticulatus).3 The tick acts as both a natural reservoir and a neither ATCC nor the U.S.