Corn Stover and Sweet Sorghum Case Studies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Educated Youth Should Go to the Rural Areas: a Tale of Education, Employment and Social Values*
Educated Youth Should Go to the Rural Areas: A Tale of Education, Employment and Social Values* Yang You† Harvard University This draft: July 2018 Abstract I use a quasi-random urban-dweller allocation in rural areas during Mao’s Mass Rustication Movement to identify human capital externalities in education, employment, and social values. First, rural residents acquired an additional 0.1-0.2 years of education from a 1% increase in the density of sent-down youth measured by the number of sent-down youth in 1969 over the population size in 1982. Second, as economic outcomes, people educated during the rustication period suffered from less non-agricultural employment in 1990. Conversely, in 2000, they enjoyed increased hiring in all non-agricultural occupations and lower unemployment. Third, sent-down youth changed the social values of rural residents who reported higher levels of trust, enhanced subjective well-being, altered trust from traditional Chinese medicine to Western medicine, and shifted job attitudes from objective cognitive assessments to affective job satisfaction. To explore the mechanism, I document that sent-down youth served as rural teachers with two new county-level datasets. Keywords: Human Capital Externality, Sent-down Youth, Rural Educational Development, Employment Dynamics, Social Values, Culture JEL: A13, N95, O15, I31, I25, I26 * This paper was previously titled and circulated, “Does living near urban dwellers make you smarter” in 2017 and “The golden era of Chinese rural education: evidence from Mao’s Mass Rustication Movement 1968-1980” in 2015. I am grateful to Richard Freeman, Edward Glaeser, Claudia Goldin, Wei Huang, Lawrence Katz, Lingsheng Meng, Nathan Nunn, Min Ouyang, Andrei Shleifer, and participants at the Harvard Economic History Lunch Seminar, Harvard Development Economics Lunch Seminar, and Harvard China Economy Seminar, for their helpful comments. -

A Simulation-Based Linear Fractional Programming Model for Adaptable Water Allocation Planning in the Main Stream of the Songhua River Basin, China
water Article A Simulation-Based Linear Fractional Programming Model for Adaptable Water Allocation Planning in the Main Stream of the Songhua River Basin, China Qiang Fu 1,2,3 ID , Linqi Li 1, Mo Li 1,* ID , Tianxiao Li 1,2, Dong Liu 1,2 and Song Cui 1 1 School of Water Conservancy & Civil Engineering, Northeast Agricultural University, Harbin 150030, China; [email protected] (Q.F.); [email protected] (L.L.); [email protected] (T.L.); [email protected] (D.L.); [email protected] (S.C.) 2 Key Laboratory of Effective Utilization of Agricultural Water Resources of Ministry of Agriculture, Northeast Agricultural University, Harbin 150030, China 3 Heilongjiang Provincial Key Laboratory of Water Resources and Water Conservancy Engineering in Cold Region, Northeast Agricultural University, Harbin 150030, China * Correspondence: [email protected]; Tel.: +86-451-5519-0209 Received: 26 April 2018; Accepted: 7 May 2018; Published: 10 May 2018 Abstract: The potential influence of natural variations in a climate system on global warming can change the hydrological cycle and threaten current strategies of water management. A simulation-based linear fractional programming (SLFP) model, which integrates a runoff simulation model (RSM) into a linear fractional programming (LFP) framework, is developed for optimal water resource planning. The SLFP model has multiple objectives such as benefit maximization and water supply minimization, balancing water conflicts among various water demand sectors, and addressing complexities of water resource allocation system. Lingo and Excel programming solutions were used to solve the model. Water resources in the main stream basin of the Songhua River are allocated for 4 water demand sectors in 8 regions during two planning periods under different scenarios. -

A Brief Introduction to the Dairy Industry in Heilongjiang NBSO Dalian
A brief introduction to the Dairy Industry in Heilongjiang NBSO Dalian RVO.nl | Brief Introduction Dairy industry Heilongjiang, NBSO Dalian Colofon This is a publication of: Netherlands Enterprise Agency Prinses Beatrixlaan 2 PO Box 93144, 2509 AC, The Hague Phone: 088 042 42 42 Email: via contact form on the website Website: www.rvo.nl This survey has been conducted by the Netherlands Business Support Office in Dalian If you have any questions regarding this business sector in Heilongjiang Province or need any form of business support, please contact NBSO Dalian: Chief Representative: Renée Derks Deputy Representative: Yin Hang Phone: +86 (0)411 3986 9998 Email: [email protected] For further information on the Netherlands Business Support Offices, see www.nbso.nl © Netherlands Enterprise Agency, August 2015 NL Enterprise Agency is a department of the Dutch ministry of Economic Affairs that implements government policy for agricultural, sustainability, innovation, and international business and cooperation. NL Enterprise Agency is the contact point for businesses, educational institutions and government bodies for information and advice, financing, networking and regulatory matters. Although a great degree of care has been taken in the preparation of this document, no rights may be derived from this brochure, or from any of the examples contained herein, nor may NL Enterprise Agency be held liable for the consequences arising from the use thereof. This publication may not be reproduced, in whole, or in part, in any form, without the prior written consent of the publisher. Page 2 of 10 RVO.nl | Brief Introduction Dairy industry Heilongjiang, NBSO Dalian Contents Colofon ..................................................................................................... -

December 1998
JANUARY - DECEMBER 1998 SOURCE OF REPORT DATE PLACE NAME ALLEGED DS EX 2y OTHER INFORMATION CRIME Hubei Daily (?) 16/02/98 04/01/98 Xiangfan C Si Liyong (34 yrs) E 1 Sentenced to death by the Xiangfan City Hubei P Intermediate People’s Court for the embezzlement of 1,700,00 Yuan (US$20,481,9). Yunnan Police news 06/01/98 Chongqing M Zhang Weijin M 1 1 Sentenced by Chongqing No. 1 Intermediate 31/03/98 People’s Court. It was reported that Zhang Sichuan Legal News Weijin murdered his wife’s lover and one of 08/05/98 the lover’s relatives. Shenzhen Legal Daily 07/01/98 Taizhou C Zhang Yu (25 yrs, teacher) M 1 Zhang Yu was convicted of the murder of his 01/01/99 Zhejiang P girlfriend by the Taizhou City Intermediate People’s Court. It was reported that he had planned to kill both himself and his girlfriend but that the police had intervened before he could kill himself. Law Periodical 19/03/98 07/01/98 Harbin C Jing Anyi (52 yrs, retired F 1 He was reported to have defrauded some 2600 Liaoshen Evening News or 08/01/98 Heilongjiang P teacher) people out of 39 million Yuan 16/03/98 (US$4,698,795), in that he loaned money at Police Weekend News high rates of interest (20%-60% per annum). 09/07/98 Southern Daily 09/01/98 08/01/98 Puning C Shen Guangyu D, G 1 1 Convicted of the murder of three children - Guangdong P Lin Leshan (f) M 1 1 reported to have put rat poison in sugar and 8 unnamed Us 8 8 oatmeal and fed it to the three children of a man with whom she had a property dispute. -
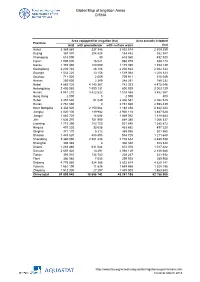
Global Map of Irrigation Areas CHINA
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52 -

Heilongjiang Road Development II Project (Yichun-Nenjiang)
Technical Assistance Consultant’s Report Project Number: TA 7117 – PRC October 2009 People’s Republic of China: Heilongjiang Road Development II Project (Yichun-Nenjiang) FINAL REPORT (Volume II of IV) Submitted by: H & J, INC. Beijing International Center, Tower 3, Suite 1707, Beijing 100026 US Headquarters: 6265 Sheridan Drive, Suite 212, Buffalo, NY 14221 In association with WINLOT No 11 An Wai Avenue, Huafu Garden B-503, Beijing 100011 This consultant’s report does not necessarily reflect the views of ADB or the Government concerned, ADB and the Government cannot be held liable for its contents. All views expressed herein may not be incorporated into the proposed project’s design. Asian Development Bank Heilongjiang Road Development II (TA 7117 – PRC) Final Report Supplementary Appendix A Financial Analysis and Projections_SF1 S App A - 1 Heilongjiang Road Development II (TA 7117 – PRC) Final Report SUPPLEMENTARY APPENDIX SF1 FINANCIAL ANALYSIS AND PROJECTIONS A. Introduction 1. Financial projections and analysis have been prepared in accordance with the 2005 edition of the Guidelines for the Financial Governance and Management of Investment Projects Financed by the Asian Development Bank. The Guidelines cover both revenue earning and non revenue earning projects. Project roads include expressways, Class I and Class II roads. All will be built by the Heilongjiang Provincial Communications Department (HPCD). When the project started it was assumed that all project roads would be revenue earning. It was then discovered that national guidance was that Class 2 roads should be toll free. The ADB agreed that the DFR should concentrate on the revenue earning Expressway and Class I roads, 2. -

Li, 2007. Selenium Deficiency and Endemic Heart Failure in China
Changsheng Li Selenium Deficiency and Endemic Heart Failure in China: A Case Study of Biogeochemistry for Human Health A PECULIAR DISEASE their research strategy, the team selected Keshan County in Heilongjiang Province, the origin of Keshan disease, as their In 1937, a terrible disease of heart failure was reported in some first study area. By teaming up with the local medical doctors, rural areas in Heilongjiang, a far northeastern province of this group conducted a thorough field survey by literally China. Women and children were its primary victims. The walking across the entire county in 1968. They visited almost all disease frequently occurred without warning and led to the the villages in the county, obtaining information on the death of a large number of people. The major symptom of the incidence of Keshan disease as well the local environmental disease was myocardial necrosis, which led to acute hypoxia, conditions. Soil and drinking water samples were collected from vomiting, and finally death in several hours. Preliminary each of the villages for chemical analysis. The investigation investigations were conducted in the late 1930s and 1940s but resulted in a map of multiyear cumulative deaths from Keshan biotic infecting agents could not be identified. The peculiar disease, with the chemical composition of the drinking water disease was then named after the county, Keshan, where the and soils at the village level described for the county. The map first cases of death from the disease were reported. Since then, demonstrated an interesting pattern of Keshan disease in its Keshan disease was found in another 12 provinces across China geographic distribution in the county. -

The Natural Control Ability of Parasitic Chalcids on Poplar Scales Quadraspidiotus Gigas (Thiem Et Gerneck) and Effect of Artificial Migration
http://www.paper.edu.cn THE NATURAL CONTROL ABILITY OF PARASITIC CHALCIDS ON POPLAR SCALES QUADRASPIDIOTUS GIGAS (THIEM ET GERNECK) AND EFFECT OF ARTIFICIAL MIGRATION Hu Yinyue Chi Defu Shao Jingwen Zhang Fengbin Li Chengde Northeast Forestry University Heilongjiang Province 150040 China E-mail [email protected] Abstract: This paper reported the investigation of the population complex of parasitic natural enemies on the main stem pests, Quadraspidiotus gigas (Thiem et Gerneck), Lepidosephes saliciana (Borchs.) and Pulvinaria salicicola (Borchs.), in protective poplar forests of Northeast China, a total of 12 species of parasitic chatcids being discovered. The species of parasitic chalcids, discovered on poplar scales were evaluated and screened using indexes of outstanding natural enemies: ecological compatibility, temporal synchronization, density responsiveness, reproductive potential, searching capacity, dispersal capacity, Spatial distribution pattern, resistance to unfavorable conditions and food requirements. Investigations discovered that Pteroptrix longiclava (Giraut) and Encarsia gigas (Tshumakova) highly synchronized with the host Q. gigas in ecological compatibility, period of growth and spatial distribution pattern, possessing high reproductive, searching and dispersal capacities, in positive density-dependent relationship in population dynamics with that of the host. The food requirement of adults of the two species was easy to satisfy. Adults were easy to preserve, with definite resistance to rain and wind. In 1992 and 1993, parasitic chalcids were artificially migrated and released in experimental forests hundreds hm2. Surveys in 1993, 1994 and 1995 showed that chalcid safely colonized these forests, average parasitic rates reaching 20.8%, 30.7% and 28.5% respectively. After two years of releasing, chalcid dispersed away for more than 30m. -

PPI Data Update Note 29 December 2009 Investment Commitments And
PPI data update note 29 December 2009 Investment commitments and the number of new projects decline in East Asia and Pacific Private activity in infrastructure in East Asia and Pacific slowed in 2008, according to just-released data from the Private Participation in Infrastructure Project Database. Both investment commitments and the number of new projects declined, in both the first and second half of the year. New energy projects and existing telecommunications operators accounted for most the annual investment. The region attracted 10% of the year’s total investment commitments in developing countries. In 2008, 78 infrastructure projects with private participation reached financial or contractual closure in 10 low- or middle-income countries of East Asia and Pacific.1 These projects involve investment commitments (hereafter, investment) of US$8.6 billion. Infrastructure projects implemented in previous years had additional commitments of US$6.8 billion, bringing total investment in 2008 to US$15.4 billion. That level was 33% lower than in 2007 and the lowest since 2002 (figure 1).2 New projects as well as existing ones accounted for the decline. When investment is classified by type, both investment in physical assets and payments to governments explain the decline. Investment in physical assets fell by 24% from the level in 2007, to US$13.8 billion in 2008 (figure 2). Payments to governments (such as concession or lease fees and divestiture revenues) dropped by 67% to US$1.6 billion. Figure 1 Investment commitments to infrastructure -

Table of Contents
TABLE OF CONTENTS 1. FALUN GONG PRACTITIONERS WHO HAVE REPORTEDLY RECEIVED PRISON SENTENCES OR ADMINISTRATIVE SENTENCES .................................................................................................................................................. 3 2. FALUN GONG PRACTITIONERS WHO MAY REMAIN IN DETENTION ................................................................... 27 3. FALUN GONG PRACTITIONERS REPORTED TO HAVE BEEN DETAINED WHOSE SUBSEQUENT FATE IS UNKNOWN ................................................................................................................................................................. 51 2 List of sentences, administrative sentences and those detained PEOPLE’S REPUBLIC OF CHINA Falun Gong practitioners: list of sentences, administrative sentences and those detained Sources : this information has been compiled from Falun Gong sources, news releases from the Information Centre for Human Rights and Democractic Movement in China, Reuters, AFP, AP and other press agencies and newspapers published prior to 18 March 2000. KEY: D = district C = city Co = county 1. FALUN GONG PRACTITIONERS WHO HAVE REPORTEDLY RECEIVED PRISON SENTENCES OR ADMINISTRATIVE SENTENCES NAME OCCUPATION PLACE OF DETENTION TRIAL/ SENTENCING CHARGE AND/OR SENTENCE NOTES ORIGIN DATE RE-EDUCAT BODY ACCUSATION ION DATE BEIJING MUNICIPALITY Ji Liewu, 36 Manager of a Hong 20/07/99 26/12/99 Beijing No.1 charged on 19/10/99 12 years’ Accused of holding a position of Kong subsidiary of a Intermediate with "illegal obtaining -

A Spatial Study on Keshan Disease Prevalence And
ORIGINAL PAPER International Journal of Occupational Medicine and Environmental Health 2021;34(5):659 – 666 https://doi.org/10.13075/ijomeh.1896.01749 A SPATIAL STUDY ON KESHAN DISEASE PREVALENCE AND SELENOPROTEIN P IN THE HEILONGJIANG PROVINCE, CHINA YANAN WANG1, XIAO ZHANG1,2, TONG WANG1, JIE HOU1, ZHONGYING GUO1,3, XIAOMIN HAN1,3, HUIHUI ZHOU1,4, HONG LIANG1,5, and ZHIFENG XING6 1 Harbin Medical University, Harbin, China Institute of Keshan Disease, Chinese Center for Endemic Disease Control 2 Qingdao Municipal Hospital, Qingdao, China Department of Healthcare-Associated Infection Management 3 Harbin Center for Disease Control and Prevention, Harbin, China Director Office 4 Jining Medical University, Jining, China School of Public Health 5 Qiqihar Medical University, Qiqihar, China School of Public Health 6 Heilongjiang Center for Disease Control and Prevention, Harbin, China Institute for Endemic Disease Control and Prevention Abstract Objectives: Few spatial studies on Keshan disease (KD) prevalence and serum selenoprotein P (SELENOP) levels have been reported in the Heilongjiang Province, China. This study aimed to investigate the spatial relationships between KD prevalence, SELENOP levels, and the socio-economic status for the pre- cise prevention and control of KD. Material and Methods: The study was carried out in all the 66 KD endemic counties in the Heilongjiang Province using a non-probability sampling method of a key village survey based on county-wide case-searching. The participants completed a questionnaire and had their serum SELENOP levels measured using enzyme-linked immunosorbent assay. Thematic maps were created, and spatial regression analysis was performed by ordinary least squares using ArcGIS 9.0. -

Molecular Characterization of the ORF3 and S1 Genes of Porcine Epidemic Diarrhea Virus Non S-INDEL Strains in Seven Regions of China, 2015
RESEARCH ARTICLE Molecular Characterization of the ORF3 and S1 Genes of Porcine Epidemic Diarrhea Virus Non S-INDEL Strains in Seven Regions of China, 2015 Enyu Wang1, Donghua Guo1, Chunqiu Li1, Shan Wei1, Zhihui Wang1, Qiujin Liu1, Bei Zhang1, Fanzhi Kong1, Li Feng2*, Dongbo Sun1* a11111 1 College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University, Sartu District, Daqing, P.R. China, 2 Division of Swine Infectious Diseases, National Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences, Nangang District, Harbin, P.R. China * [email protected] (DS); [email protected] (LF) OPEN ACCESS Abstract Citation: Wang E, Guo D, Li C, Wei S, Wang Z, Liu Q, et al. (2016) Molecular Characterization of the In an effort to trace the evolution of porcine epidemic diarrhea virus (PEDV), S1 and ORF3 ORF3 and S1 Genes of Porcine Epidemic Diarrhea genes of viruses identified in 41 pig farms from seven regions (North, Northeast, Northwest, Virus Non S-INDEL Strains in Seven Regions of Central, East, South West, and South, respectively) of China in 2015 were sequenced and China, 2015. PLoS ONE 11(8): e0160561. ORF3 S1 doi:10.1371/journal.pone.0160561 analyzed. Sequence analysis revealed that the 41 genes and 29 genes identified in our study exhibited nucleotide homologies of 98.2%–100% and 96.6%–100%, respectively; Editor: Zhefeng Meng, Fudan University, CHINA these two genes exhibited low nucleotide sequence similarities with classical CV777 strain Received: March 23, 2016 and early Chinese strain LZC. Phylogenetic analysis indicated that the identified PEDV Accepted: July 21, 2016 strains belonged to global non S-INDEL strains, and exhibited genetic diversity; S1 gene of 1130 Published: August 5, 2016 the HLJ2015/DP1-1 strain harbored an unique deletion of 12 nucleotides (A CAACTCCA 1141 Copyright: © 2016 Wang et al.