CAVASS: a Computer Assisted Visualization and Analysis Software
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours
A 3D interactive multi-object segmentation tool using local robust statistics driven active contours The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Gao, Yi, Ron Kikinis, Sylvain Bouix, Martha Shenton, and Allen Tannenbaum. 2012. A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours. Medical Image Analysis 16, no. 6: 1216–1227. doi:10.1016/j.media.2012.06.002. Published Version doi:10.1016/j.media.2012.06.002 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:28548930 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA NIH Public Access Author Manuscript Med Image Anal. Author manuscript; available in PMC 2013 August 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Med Image Anal. 2012 August ; 16(6): 1216–1227. doi:10.1016/j.media.2012.06.002. A 3D Interactive Multi-object Segmentation Tool using Local Robust Statistics Driven Active Contours Yi Gaoa,*, Ron Kikinisb, Sylvain Bouixa, Martha Shentona, and Allen Tannenbaumc aPsychiatry Neuroimaging Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 bSurgical Planning Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 cDepartments of Electrical and Computer Engineering and Biomedical Engineering, Boston University, Boston, MA 02115 Abstract Extracting anatomical and functional significant structures renders one of the important tasks for both the theoretical study of the medical image analysis, and the clinical and practical community. -

Road Lane Detection for Android
MASARYK UNIVERSITY FACULTY}w¡¢£¤¥¦§¨ OF I !"#$%&'()+,-./012345<yA|NFORMATICS Road lane detection for Android BACHELOR THESIS Jakub Medvecký-Heretik Brno, spring 2014 Declaration I do hereby declare, that this paper is my original authorial work, which I have worked on by my own. All sources, references and liter- ature used or excerpted during elaboration of this work are properly cited and listed in complete reference to the due source. Jakub Medvecký-Heretik Advisor: Mgr. Dušan Klinec Acknowledgement I would like to thank my supervisor, Mgr. Dušan Klinec for his con- tinual support and advice throughout this work. I would also like to thank my family and friends for their support. Abstract The aim of this bachelor thesis was to analyze the possibilities of im- age processing techniques on the Android mobile platform for the purposes of a computer vision application focusing on the problem of road lane detection. Utilizing the results of the analysis an appli- cation was created, which detects lines and circles in an image in real time using various approaches to segmentation and different imple- mentations of the Hough transform. The application makes use of the OpenCV library and the results achieved with its help are com- pared to the results achieved with my own implementation. Keywords computer vision, road lane detection, circle detection, image process- ing, segmentation, Android, OpenCV, Hough transform Contents 1 Introduction ............................ 3 2 Computer vision ......................... 5 2.1 Image processing ...................... 5 2.2 Techniques .......................... 6 2.3 Existing libraries ...................... 6 2.3.1 OpenCV . 6 2.3.2 VXL . 7 2.3.3 ImageJ . -

Oracle FLEXCUBE Core Banking Licensing Guide Release 11.5.0.0.0
Oracle FLEXCUBE Core Banking Licensing Guide Release 11.5.0.0.0 Part No. E52876-01 July 2014 Licensing Guide July 2014 Oracle Financial Services Software Limited Oracle Park Off Western Express Highway Goregaon (East) Mumbai, Maharashtra 400 063 India Worldwide Inquiries: Phone: +91 22 6718 3000 Fax:+91 22 6718 3001 www.oracle.com/financialservices/ Copyright © 2008, 2014, Oracle and/or its affiliates. All rights reserved. Oracle and Java are registered trademarks of Oracle and/or its affiliates. Other names may be trademarks of their respective owners. U.S. GOVERNMENT END USERS: Oracle programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, delivered to U.S. Government end users are “commercial computer software” pursuant to the applicable Federal Acquisition Regulation and agency-specific supplemental regulations. As such, use, duplication, disclosure, modification, and adaptation of the programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, shall be subject to license terms and license restrictions applicable to the programs. No other rights are granted to the U.S. Government. This software or hardware is developed for general use in a variety of information management applications. It is not developed or intended for use in any inherently dangerous applications, including applications that may create a risk of personal injury. If you use this software or hardware in dangerous applications, then you shall be responsible to take all appropriate failsafe, backup, redundancy, and other measures to ensure its safe use. Oracle Corporation and its affiliates disclaim any liability for any damages caused by use of this software or hardware in dangerous applications. -

Java Advanced Imaging
Java Advanced Imaging MIMUC Medientechnik SS 2003 Übersicht Wozu Bildbearbeitung mit Frameworks? Warum JAI? Verzahnung mit Java2D (AWT) JAI Packages und Typen Unterstützte Codecs und Bildformate BufferedImage, Raster, ColorModel (AWT) Rendergraphen Aufbau der Bildklassen Anzeigen eines gespeicherten Bildes Parameterblock Operationen Look-Up-Table Speichern eines Bildes Quellangabe MIMUC Medientechnik SS 2003 Helge Groß 2 - 23 Wozu Bildbearbeitung mit Frameworks? Rechner/Monitore können Bilder heute schnell verarbeiten/anzeigen Dadurch ist Analyse- und Bearbeitungsmöglichkeit am PC geschaffen Frameworks stellen bereit Bildmanipulationsmöglichkeiten Mathematische Operationen Bildanzeigemöglichkeit Bild-Codecs Einsatzgebiete Bildanalyse- und Bildanzeigesysteme Objektpositionierung, Objekttracking, Qualitätsprüfung Bildbearbeitungssysteme Medien, Werbeindustrie, Grafik-Design, Zeichenprogramme Applikationen Programme, Computerspiele World Wide Web Webseiten MIMUC Medientechnik SS 2003 Helge Groß 3 - 23 Warum JAI? Pro Plattformunabhängig Geringer Speicherplatz Schnelle Internetübertragung Applets im WWW Bildverarbeitende Web-Applikationen Als Frontend einer anderen Programmiersprache Diese übernimmt komplexere Operationen Client-Server-Architektur Komplexe Bildmanipulationen durchaus praktikabel Großer Leistungsumfang zur Bildverarbeitung Versucht alle Bedürfnisse zu erfüllen Extrem Erweiterbar Contra Langsam nicht zuletzt wegen Swing MIMUC Medientechnik SS 2003 Helge Groß 4 - 23 Verzahnung mit Java2D (AWT) JAI -

An Open-Source Research Platform for Image-Guided Therapy
Int J CARS DOI 10.1007/s11548-015-1292-0 ORIGINAL ARTICLE CustusX: an open-source research platform for image-guided therapy Christian Askeland1,3 · Ole Vegard Solberg1 · Janne Beate Lervik Bakeng1 · Ingerid Reinertsen1 · Geir Arne Tangen1 · Erlend Fagertun Hofstad1 · Daniel Høyer Iversen1,2,3 · Cecilie Våpenstad1,2 · Tormod Selbekk1,3 · Thomas Langø1,3 · Toril A. Nagelhus Hernes2,3 · Håkon Olav Leira2,3 · Geirmund Unsgård2,3 · Frank Lindseth1,2,3 Received: 3 July 2015 / Accepted: 31 August 2015 © The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Results The validation experiments show a navigation sys- Purpose CustusX is an image-guided therapy (IGT) research tem accuracy of <1.1mm, a frame rate of 20 fps, and latency platform dedicated to intraoperative navigation and ultra- of 285ms for a typical setup. The current platform is exten- sound imaging. In this paper, we present CustusX as a robust, sible, user-friendly and has a streamlined architecture and accurate, and extensible platform with full access to data and quality process. CustusX has successfully been used for algorithms and show examples of application in technologi- IGT research in neurosurgery, laparoscopic surgery, vascular cal and clinical IGT research. surgery, and bronchoscopy. Methods CustusX has been developed continuously for Conclusions CustusX is now a mature research platform more than 15years based on requirements from clinical for intraoperative navigation and ultrasound imaging and is and technological researchers within the framework of a ready for use by the IGT research community. CustusX is well-defined software quality process. The platform was open-source and freely available at http://www.custusx.org. -

Medical Image Processing Software
Wohlers Report 2018 Medical Image Processing Software Medical image Patient-specific medical devices and anatomical models are almost always produced using radiological imaging data. Medical image processing processing software is used to translate between radiology file formats and various software AM file formats. Theoretically, any volumetric radiological imaging dataset by Andy Christensen could be used to create these devices and models. However, without high- and Nicole Wake quality medical image data, the output from AM can be less than ideal. In this field, the old adage of “garbage in, garbage out” definitely applies. Due to the relative ease of image post-processing, computed tomography (CT) is the usual method for imaging bone structures and contrast- enhanced vasculature. In the dental field and for oral- and maxillofacial surgery, in-office cone-beam computed tomography (CBCT) has become popular. Another popular imaging technique that can be used to create anatomical models is magnetic resonance imaging (MRI). MRI is less useful for bone imaging, but its excellent soft tissue contrast makes it useful for soft tissue structures, solid organs, and cancerous lesions. Computed tomography: CT uses many X-ray projections through a subject to computationally reconstruct a cross-sectional image. As with traditional 2D X-ray imaging, a narrow X-ray beam is directed to pass through the subject and project onto an opposing detector. To create a cross-sectional image, the X-ray source and detector rotate around a stationary subject and acquire images at a number of angles. An image of the cross-section is then computed from these projections in a post-processing step. -
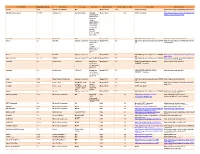
Third Party Version
Third Party Name Third Party Version Manufacturer License Type Comments Merge Product Merge Product Versions License details Software source autofac 3.5.2 Autofac Contributors MIT Merge Cardio 10.2 SOUP repository https://www.nuget.org/packages/Autofac/3.5 .2 Gibraltar Loupe Agent 2.5.2.815 eSymmetrix Gibraltor EULA Gibraltar Merge Cardio 10.2 SOUP repository https://my.gibraltarsoftware.com/Support/Gi Loupe Agent braltar_2_5_2_815_Download will be used within the Cardio Application to view events and metrics so you can resolve support issues quickly and easily. Modernizr 2.8.3 Modernizr MIT Merge Cadio 6.0 http://modernizr.com/license/ http://modernizr.com/download/ drools 2.1 Red Hat Apache License 2.0 it is a very old Merge PACS 7.0 http://www.apache.org/licenses/LICENSE- http://mvnrepository.com/artifact/drools/dro version of 2.0 ols-spring/2.1 drools. Current version is 6.2 and license type is changed too drools 6.3 Red Hat Apache License 2.0 Merge PACS 7.1 http://www.apache.org/licenses/LICENSE- https://github.com/droolsjbpm/drools/releases/ta 2.0 g/6.3.0.Final HornetQ 2.2.13 v2.2..13 JBOSS Apache License 2.0 part of JBOSS Merge PACS 7.0 http://www.apache.org/licenses/LICENSE- http://mvnrepository.com/artifact/org.hornet 2.0 q/hornetq-core/2.2.13.Final jcalendar 1.0 toedter.com LGPL v2.1 MergePacs Merge PACS 7.0 GNU LESSER GENERAL PUBLIC http://toedter.com/jcalendar/ server uses LICENSE Version 2. v1, and viewer uses v1.3. -

André Vilas Boas Da Costa Retscan: Efficient Fovea and Optic Disc Detection in Retinographies
Universidade do Minho Escola de Engenharia André Vilas Boas da Costa RetScan: Efficient Fovea and Optic Disc Detection in Retinographies Outubro de 2011 Universidade do Minho Escola de Engenharia Departamento de Informática André Vilas Boas da Costa RetScan: Efficient Fovea and Optic Disc Detection in Retinographies Dissertação de Mestrado Mestrado de Informática Trabalho realizado sob orientação de Alberto José Proença Outubro de 2011 Abstract The Fovea and Optic Disc are relevant anatomical eye structures to diagnose var- ious diseases. Its automatic detection can provide both a cost reduction when analysing large populations and improve the effectiveness of ophthalmologists and optometrists. This dissertation describes a methodology to automatically detect these structures and analyses a, CPU only, MATLAB implementation of this methodology. RetScan is a port to a freeware environment of this methodology, its functionality and perfor- mance are evaluated and compared to the original. The results of both evaluations lead to a discussion on possible improvements in the metodology that influence the functionality and performance. The resulting improvements are implemented and integrated in RetScan. To further improve performance, a parallelization of RetScan to take advantage of a multi-core architecture or a CUDA-enabled accelerator was designed, coded and evaluated. This evaluation reveals that RetScan achieves its best throughput efficiency using a multi-core architecture only and analysing several images at once. For one image usage, using multi-core only is also the best solution, but with a small speed-up. The usage of CUDA-enabled accelerators is not recom- mended for this scope as the images are small and the cost of the data transfer to and from the accelerator has a severe impact on performance. -

An Open Source Freeware Software for Ultrasound Imaging and Elastography
Proceedings of the eNTERFACE’07 Workshop on Multimodal Interfaces, Istanbul,˙ Turkey, July 16 - August 10, 2007 USIMAGTOOL: AN OPEN SOURCE FREEWARE SOFTWARE FOR ULTRASOUND IMAGING AND ELASTOGRAPHY Ruben´ Cardenes-Almeida´ 1, Antonio Tristan-Vega´ 1, Gonzalo Vegas-Sanchez-Ferrero´ 1, Santiago Aja-Fernandez´ 1, Veronica´ Garc´ıa-Perez´ 1, Emma Munoz-Moreno˜ 1, Rodrigo de Luis-Garc´ıa 1, Javier Gonzalez-Fern´ andez´ 2, Dar´ıo Sosa-Cabrera 2, Karl Krissian 2, Suzanne Kieffer 3 1 LPI, University of Valladolid, Spain 2 CTM, University of Las Palmas de Gran Canaria 3 TELE Laboratory, Universite´ catholique de Louvain, Louvain-la-Neuve, Belgium ABSTRACT • Open source code: to be able for everyone to modify and reuse the source code. UsimagTool will prepare specific software for the physician to change parameters for filtering and visualization in Ultrasound • Efficiency, robust and fast: using a standard object ori- Medical Imaging in general and in Elastography in particular, ented language such as C++. being the first software tool for researchers and physicians to • Modularity and flexibility for developers: in order to chan- compute elastography with integrated algorithms and modular ge or add functionalities as fast as possible. coding capabilities. It will be ready to implement in different • Multi-platform: able to run in many Operating systems ecographic systems. UsimagTool is based on C++, and VTK/ITK to be useful for more people. functions through a hidden layer, which means that participants may import their own functions and/or use the VTK/ITK func- • Usability: provided with an easy to use GUI to interact tions. as easy as possible with the end user. -

Programming in Java Advanced Imaging
Programming in Java™ Advanced Imaging Release 1.0.1 November 1999 JavaSoft A Sun Microsystems, Inc. Business 901 San Antonio Road Palo Alto, CA 94303 USA 415 960-1300 fax 415 969-9131 1999 Sun Microsystems, Inc. 901 San Antonio Road, Palo Alto, California 94303 U.S.A. All rights reserved. RESTRICTED RIGHTS LEGEND: Use, duplication, or disclosure by the United States Government is subject to the restrictions set forth in DFARS 252.227-7013 (c)(1)(ii) and FAR 52.227-19. The release described in this document may be protected by one or more U.S. patents, for- eign patents, or pending applications. Sun Microsystems, Inc. (SUN) hereby grants to you a fully paid, nonexclusive, nontrans- ferable, perpetual, worldwide limited license (without the right to sublicense) under SUN’s intellectual property rights that are essential to practice this specification. This license allows and is limited to the creation and distribution of clean-room implementa- tions of this specification that (i) are complete implementations of this specification, (ii) pass all test suites relating to this specification that are available from SUN, (iii) do not derive from SUN source code or binary materials, and (iv) do not include any SUN binary materials without an appropriate and separate license from SUN. Java, JavaScript, Java 3D, and Java Advanced Imaging are trademarks of Sun Microsys- tems, Inc. Sun, Sun Microsystems, the Sun logo, Java and HotJava are trademarks or reg- istered trademarks of Sun Microsystems, Inc. UNIX® is a registered trademark in the United States and other countries, exclusively licensed through X/Open Company, Ltd. -

CES Free Or Open Source Licenses Licenses Library Version 15.2 2020R1
CES Free or Open Source Licenses Licenses Library Version 15.2 2020R1 Revision 2.00 December 2020 Verint.com Twitter.com/verint Facebook.com/verint Blog.verint.com Table of Contents Free or Open Source Licenses ....................................................................................... 1 7-Zip - GNU LGPL + unRAR restrictions .................................................................... 1 ActivePython ............................................................................................................... 2 ANTLR .......................................................................................................................... 6 Apache License............................................................................................................ 6 ares Library................................................................................................................. 11 Attribution-NonCommercial-ShareAlike 3.0 Unported ............................................. 12 Batik SVG Toolkit ....................................................................................................... 17 Bouncy Castle ............................................................................................................ 19 Boost ........................................................................................................................... 20 BSD (4-Clause) License ............................................................................................ 20 COMMON DEVELOPMENT AND DISTRIBUTION