Engineering Hepadnaviruses As Reporter-Expressing Vectors: Recent Progress and Future Perspectives
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hepatitis Virus in Long-Fingered Bats, Myanmar
DISPATCHES Myanmar; the counties are adjacent to Yunnan Province, Hepatitis Virus People’s Republic of China. The bats covered 6 species: Miniopterus fuliginosus (n = 640), Hipposideros armiger in Long-Fingered (n = 8), Rhinolophus ferrumequinum (n = 176), Myotis chi- nensis (n = 11), Megaderma lyra (n = 6), and Hipposideros Bats, Myanmar fulvus (n = 12). All bat tissue samples were subjected to vi- Biao He,1 Quanshui Fan,1 Fanli Yang, ral metagenomic analysis (unpublished data). The sampling Tingsong Hu, Wei Qiu, Ye Feng, Zuosheng Li, of bats for this study was approved by the Administrative Yingying Li, Fuqiang Zhang, Huancheng Guo, Committee on Animal Welfare of the Institute of Military Xiaohuan Zou, and Changchun Tu Veterinary, Academy of Military Medical Sciences, China. We used PCR to further study the prevalence of or- During an analysis of the virome of bats from Myanmar, thohepadnavirus in the 6 bat species; the condition of the a large number of reads were annotated to orthohepadnavi- samples made serologic assay and pathology impracticable. ruses. We present the full genome sequence and a morpho- Viral DNA was extracted from liver tissue of each of the logical analysis of an orthohepadnavirus circulating in bats. 853 bats by using the QIAamp DNA Mini Kit (QIAGEN, This virus is substantially different from currently known Hilden, Germany). To detect virus in the samples, we con- members of the genus Orthohepadnavirus and represents ducted PCR by using the TaKaRa PCR Kit (TaKaRa, Da- a new species. lian, China) with a pair of degenerate pan-orthohepadnavi- rus primers (sequences available upon request). The PCR he family Hepadnaviridae comprises 2 genera (Ortho- reaction was as follows: 45 cycles of denaturation at 94°C Thepadnavirus and Avihepadnavirus), and viruses clas- for 30 s, annealing at 54°C for 30 s, extension at 72°C for sified within these genera have a narrow host range. -

Virus Particle Structures
Virus Particle Structures Virus Particle Structures Palmenberg, A.C. and Sgro, J.-Y. COLOR PLATE LEGENDS These color plates depict the relative sizes and comparative virion structures of multiple types of viruses. The renderings are based on data from published atomic coordinates as determined by X-ray crystallography. The international online repository for 3D coordinates is the Protein Databank (www.rcsb.org/pdb/), maintained by the Research Collaboratory for Structural Bioinformatics (RCSB). The VIPER web site (mmtsb.scripps.edu/viper), maintains a parallel collection of PDB coordinates for icosahedral viruses and additionally offers a version of each data file permuted into the same relative 3D orientation (Reddy, V., Natarajan, P., Okerberg, B., Li, K., Damodaran, K., Morton, R., Brooks, C. and Johnson, J. (2001). J. Virol., 75, 11943-11947). VIPER also contains an excellent repository of instructional materials pertaining to icosahedral symmetry and viral structures. All images presented here, except for the filamentous viruses, used the standard VIPER orientation along the icosahedral 2-fold axis. With the exception of Plate 3 as described below, these images were generated from their atomic coordinates using a novel radial depth-cue colorization technique and the program Rasmol (Sayle, R.A., Milner-White, E.J. (1995). RASMOL: biomolecular graphics for all. Trends Biochem Sci., 20, 374-376). First, the Temperature Factor column for every atom in a PDB coordinate file was edited to record a measure of the radial distance from the virion center. The files were rendered using the Rasmol spacefill menu, with specular and shadow options according to the Van de Waals radius of each atom. -

Discovery of a Highly Divergent Hepadnavirus in Shrews from China
Virology 531 (2019) 162–170 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/virology Discovery of a highly divergent hepadnavirus in shrews from China T Fang-Yuan Niea,b,1, Jun-Hua Tianc,1, Xian-Dan Lind,1, Bin Yuc, Jian-Guang Xinge, Jian-Hai Caof, ⁎ ⁎⁎ Edward C. Holmesg,h, Runlin Z. Maa, , Yong-Zhen Zhangb,h, a State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China b State Key Laboratory for Infectious Disease Prevention and Control; Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases; Department of Zoonoses, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing, China c Wuhan Center for Disease Control and Prevention, Wuhan, China d Wenzhou Center for Disease Control and Prevention, Wenzhou, Zhejiang Province, China e Wencheng Center for Disease Control and Prevention, Wencheng, Zhejiang Province, China f Longwan Center for Disease Control and Prevention, Longwan District, Wenzhou, Zhejiang Province, China g Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life and Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia h Shanghai Public Health Clinical Center & Institute of Biomedical Sciences, Fudan University, Shanghai, China ARTICLE INFO ABSTRACT Keywords: Limited sampling means that relatively little is known about the diversity and evolutionary history of mam- Hepadnaviruses malian members of the Hepadnaviridae (genus Orthohepadnavirus). An important case in point are shrews, the Shrews fourth largest group of mammals, but for which there is limited knowledge on the role they play in viral evo- Phylogeny lution and emergence. -

And Giant Guitarfish (Rhynchobatus Djiddensis)
VIRAL DISCOVERY IN BLUEGILL SUNFISH (LEPOMIS MACROCHIRUS) AND GIANT GUITARFISH (RHYNCHOBATUS DJIDDENSIS) BY HISTOPATHOLOGY EVALUATION, METAGENOMIC ANALYSIS AND NEXT GENERATION SEQUENCING by JENNIFER ANNE DILL (Under the Direction of Alvin Camus) ABSTRACT The rapid growth of aquaculture production and international trade in live fish has led to the emergence of many new diseases. The introduction of novel disease agents can result in significant economic losses, as well as threats to vulnerable wild fish populations. Losses are often exacerbated by a lack of agent identification, delay in the development of diagnostic tools and poor knowledge of host range and susceptibility. Examples in bluegill sunfish (Lepomis macrochirus) and the giant guitarfish (Rhynchobatus djiddensis) will be discussed here. Bluegill are popular freshwater game fish, native to eastern North America, living in shallow lakes, ponds, and slow moving waterways. Bluegill experiencing epizootics of proliferative lip and skin lesions, characterized by epidermal hyperplasia, papillomas, and rarely squamous cell carcinoma, were investigated in two isolated poopulations. Next generation genomic sequencing revealed partial DNA sequences of an endogenous retrovirus and the entire circular genome of a novel hepadnavirus. Giant Guitarfish, a rajiform elasmobranch listed as ‘vulnerable’ on the IUCN Red List, are found in the tropical Western Indian Ocean. Proliferative skin lesions were observed on the ventrum and caudal fin of a juvenile male quarantined at a public aquarium following international shipment. Histologically, lesions consisted of papillomatous epidermal hyperplasia with myriad large, amphophilic, intranuclear inclusions. Deep sequencing and metagenomic analysis produced the complete genomes of two novel DNA viruses, a typical polyomavirus and a second unclassified virus with a 20 kb genome tentatively named Colossomavirus. -

Molecular Characterization of Human Papillomavirus Type 159 (HPV159)
viruses Article Molecular Characterization of Human Papillomavirus Type 159 (HPV159) Iva Markovi´c 1, Lea Hošnjak 2 , Katja Seme 2 and Mario Poljak 2,* 1 Division of Molecular Biology, Ruder¯ Boškovi´cInstitute, BijeniˇckaCesta 54, 10000 Zagreb, Croatia; [email protected] 2 Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, Zaloška Cesta 4, 1105 Ljubljana, Slovenia; [email protected] (L.H.); [email protected] (K.S.) * Correspondence: [email protected]; Tel.: +386-1-543-7453 Abstract: Human papillomavirus type 159 (HPV159) was identified in an anal swab sample and preliminarily genetically characterized by our group in 2012. Here we present a detailed molecular in silico analysis that showed that the HPV159 viral genome is 7443 bp in length and divided into five early and two late genes, with conserved functional domains and motifs, and a non-coding long control region (LCR) with significant regulatory sequences that allow the virus to complete its life cycle and infect novel host cells. HPV159, clustering into the cutaneotropic Betapapillomavirus (Beta-PV) genus, is phylogenetically most similar to HPV9, forming an individual phylogenetic group in the viral species Beta-2. After testing a large representative collection of clinical samples with HPV159 type-specific RT-PCR, in addition to the anal canal from which the first HPV159 isolate was obtained, HPV159 was further detected in other muco-cutaneous (4/181, 2.2%), mucosal (22/764, 2.9%), and cutaneous (14/554, 2.5%) clinical samples, suggesting its extensive tissue tropism. However, because very low HPV159 viral loads were estimated in the majority of positive samples, Citation: Markovi´c,I.; Hošnjak, L.; it seemed that HPV159 mainly caused clinically insignificant infections of the skin and mucosa. -
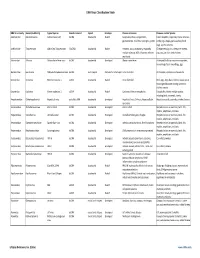
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

In Vitro Selection and Characterization of Single Stranded DNA Aptamers Inhibiting the Hepatitis B Virus Capsid-Envelope Interaction
Aus dem Veterinärwissenschaftlichen Department der Tierärztlichen Fakultät der Ludwig-Maximilians-Universität München Arbeit angefertigt unter der Leitung von: Univ.-Prof. Dr. Gerd Sutter Angefertigt im Institut für Virologie des Helmholtz Zentrum München (apl.-Prof. Dr. Volker Bruss) In vitro Selection and Characterization of single stranded DNA Aptamers Inhibiting the Hepatitis B Virus Capsid-Envelope Interaction Inaugural-Dissertation zur Erlangung der tiermedizinischen Doktorwürde der Tierärztlichen Fakultät der Ludwig-Maximilians-Universität München von Ahmed El-Sayed Abd El-Halem Orabi aus Sharkia/Ägypten München 2013 Gedruckt mit der Genehmigung der Tierärztlichen Fakultät der Ludwig-Maximilians-Universität München Dekan: Univ.-Prof. Dr. Joachim Braun Berichterstatter: Univ.-Prof. Dr. Gerd Sutter Korreferent: Univ.-Prof. Dr. Bernd Kaspers Tag der Promotion: 20. Juli 2013 My Family Contents Contents 1 INTRODUCTION ................................................................................................... 1 2 REVIEW OF THE LITERATURE ....................................................................... 2 2.1 HEPATITIS B VIRUS (HBV) .............................................................................................. 2 2.1.1 HISTORY AND TAXONOMY .......................................................................................................... 2 2.1.2 EPIDEMIOLOGY AND PATHOGENESIS .......................................................................................... 3 2.1.3 VIRION STRUCTURE .................................................................................................................... -

The Approved List of Biological Agents Advisory Committee on Dangerous Pathogens Health and Safety Executive
The Approved List of biological agents Advisory Committee on Dangerous Pathogens Health and Safety Executive © Crown copyright 2021 First published 2000 Second edition 2004 Third edition 2013 Fourth edition 2021 You may reuse this information (excluding logos) free of charge in any format or medium, under the terms of the Open Government Licence. To view the licence visit www.nationalarchives.gov.uk/doc/ open-government-licence/, write to the Information Policy Team, The National Archives, Kew, London TW9 4DU, or email [email protected]. Some images and illustrations may not be owned by the Crown so cannot be reproduced without permission of the copyright owner. Enquiries should be sent to [email protected]. The Control of Substances Hazardous to Health Regulations 2002 refer to an ‘approved classification of a biological agent’, which means the classification of that agent approved by the Health and Safety Executive (HSE). This list is approved by HSE for that purpose. This edition of the Approved List has effect from 12 July 2021. On that date the previous edition of the list approved by the Health and Safety Executive on the 1 July 2013 will cease to have effect. This list will be reviewed periodically, the next review is due in February 2022. The Advisory Committee on Dangerous Pathogens (ACDP) prepares the Approved List included in this publication. ACDP advises HSE, and Ministers for the Department of Health and Social Care and the Department for the Environment, Food & Rural Affairs and their counterparts under devolution in Scotland, Wales & Northern Ireland, as required, on all aspects of hazards and risks to workers and others from exposure to pathogens. -

Genetic Analysis of Bipartite Geminivirus Tissue Tropism
Virology 291, 311–323 (2001) doi:10.1006/viro.2001.1205, available online at http://www.idealibrary.com on View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Genetic Analysis of Bipartite Geminivirus Tissue Tropism Ying Qin1 and Ian T. D. Petty2 Department of Microbiology, North Carolina State University, Raleigh, North Carolina 27695-7615 Received July 31, 2001; returned to author for revision September 3, 2001; accepted September 18, 2001 The bipartite geminiviruses bean golden mosaic virus (BGMV), cabbage leaf curl virus (CabLCV), and tomato golden mosaic virus (TGMV) exhibit differential tissue tropism in Nicotiana benthamiana. In systemically infected leaves, BGMV remains largely confined to vascular-associated cells (phloem-limited), whereas CabLCV and TGMV can escape into the surrounding mesophyll. Previous work established that TGMV BRi, the noncoding region upstream from the BR1 open reading frame (ORF), is required for mesophyll invasion, but the virus must also contain the TGMV AL23 or BL1/BR1 ORFs. Here we show that, in a BGMV-based hybrid virus, CabLCV AL23 also directed efficient mesophyll invasion in conjunction with TGMV BRi, which suggests that host-adaptation of AL23 is important for the phenotype. Cis-acting elements required for mesophyll invasion were delineated by analyzing BGMV-based hybrid viruses in which various parts of BRi were exchanged with those of TGMV. Interestingly, mesophyll invasion efficiency of hybrid viruses was not correlated with the extent of viral DNA accumulation. In conjunction with TGMV AL23, a 52-bp region of TGMV BRi with sequence homology to DNA A was sufficient for mesophyll invasion. -

Viral Vectors 101 a Desktop Resource
Viral Vectors 101 A Desktop Resource Created and Compiled by Addgene www.addgene.org August 2018 (1st Edition) Viral Vectors 101: A Desktop Resource (1st Edition) Viral Vectors 101: A desktop resource This page intentionally left blank. 2 Chapter 1 - What Are Fluorescent Proteins? ViralViral Vectors Vector 101: A Desktop Resource (1st Edition) ViralTHE VectorsHISTORY 101: OFIntroduction FLUORESCENT to this desktop PROTEINS resource (CONT’D)By Tyler J. Ford | July 16, 2018 Dear Reader, If you’ve worked with mammalian cells, it’s likely that you’ve worked with viral vectors. Viral vectors are engineered forms of mammalian viruses that make use of natural viral gene delivery machineries and that are optimized for safety and delivery. These incredibly useful tools enable you to easily deliver genes to mammalian cells and to control gene expression in a variety of ways. Addgene has been distributing viral vectors since nearly its inception in 2004. Since then, our viral Cummulative ready-to-use virus distribution through June 2018. vector collection has grown to include retroviral vectors, lentiviral vectors, adenoviral vectors, and adeno-associated viral vectors. To further enable researchers, we started our viral service in 2017. Through this service, we distribute ready-to- use, quality-controlled AAV and lentivirus for direct use in experiments. As you can see in the chart to the left, this service is already very popular and its use has grown exponentially. With this Viral Vectors 101 eBook, we are proud to further expand our viral vector offerings. Within it, you’ll find nearly all of our viral vector educational content in a single downloadable resource. -

Orthohepadnavirus Avihepadnavirus
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2016.019aD officers) Short title: One new species in the genus Orthohepadnavirus (e.g. 6 new species in the genus Zetavirus) Modules attached 2 3 4 5 (modules 1 and 10 are required) 6 7 8 9 10 Author(s): Heléne Norder and Lars Magnius Corresponding author with e-mail address: [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . If Hepadnaviridae & Hepatitis delta virus Study in doubt, contact the appropriate subcommittee chair (fungal, invertebrate, plant, prokaryote or Group vertebrate viruses) ICTV Study Group comments (if any) and response of the proposer: Date first submitted to ICTV: July 12, 2016 Date of this revision (if different to above): August 2, 2016 ICTV-EC comments and response of the proposer: Decision: Response: Page 1 of 5 MODULE 2: NEW SPECIES creating and naming one or more new species. If more than one, they should be a group of related species belonging to the same genus. All new species must be placed in a higher taxon. -

New Avian Hepadnavirus in Palaeognathous Bird, Germany
RESEARCH LETTERS New Avian Hepadnavirus includes emus (Dromaius novaehollandiae) and ostrich- es (Struthio spp.). in Palaeognathous Bird, In 2015, a deceased adult elegant-crested tinamou Germany kept at Wuppertal Zoo (Wuppertal, Germany) underwent necropsy at the University of Veterinary Medicine Han- 1 1 nover, Foundation (Hannover, Germany). Initial histologic Wendy K. Jo, Vanessa M. Pfankuche, examination revealed moderate, necrotizing hepatitis and Henning Petersen, Samuel Frei, Maya Kummrow, inclusion body–like structures within the hepatocytes. To Stephan Lorenzen, Martin Ludlow, Julia Metzger, identify a putative causative agent, we isolated nucleic ac- Wolfgang Baumgärtner, Albert Osterhaus, ids from the liver and prepared them for sequencing on an Erhard van der Vries Illumina MiSeq system (Illumina, San Diego, CA, USA) Author affiliations: University of Veterinary Medicine Hannover, (online Technical Appendix, https://wwwnc.cdc.gov/EID/ Foundation, Hannover, Germany (W.K. Jo, V.M. Pfankuche, article/23/12/16-1634-Techapp1.pdf). We compared ob- H. Petersen, M. Ludlow, J. Metzger, W. Baumgärtner, tained reads with sequences in GenBank using an in-house A. Osterhaus, E. van der Vries); Center for Systems metagenomics pipeline. Approximately 78% of the reads Neuroscience, Hannover (W.K. Jo, V.M. Pfankuche, aligned to existing avihepadnavirus sequences. A full ge- W. Baumgärtner, A. Osterhaus); Wuppertal Zoo, Wuppertal, nome (3,024 bp) of the putative elegant-crested tinamou Germany (S. Frei, M. Kummrow); Bernhard Nocht Institute for HBV (ETHBV) was subsequently constructed by de novo Tropical Medicine, Hamburg (S. Lorenzen); Artemis One Health, assembly mapping >2 million reads (88.6%) to the virus Utrecht, the Netherlands (A. Osterhaus) genome (GenBank accession no.