Genetic Variants in Epigenetic Pathways and Risks of Multiple Cancers in the GAME-ON Consortium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rabbit Anti-SEN1/FITC Conjugated Antibody
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-SEN1/FITC Conjugated antibody SL7576R-FITC Product Name: Anti-SEN1/FITC Chinese Name: FITC标记的细胞衰老相关蛋白1抗体 Cell senescence related gene complementation group B; Cellular senescence related protein 1; CSR; CSRB; MORF 4; MORF4; MORF4 protein; Mortality factor 4; SEN 1; Alias: SEN; Senescence (cellular) related 1; Senescence related cellular 1; Transcription factor like protein MORF4; MO4L1_HUMAN; Mortality factor 4-like protein 1; MORF- related gene 15 protein; Protein MSL3-1; Transcription factor-like protein MRG15. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Dog,Cow,Zebrafish, IF=1:50-200 Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 27kDa Form: Lyophilized or Liquid Concentration: 1mg/ml immunogen: KLH conjugated synthetic peptide derived from human Mortality factor 4-like protein 1 Lsotype: IgGwww.sunlongbiotech.com Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. background: Cellular senescence, the terminal nondividing state that normal cells enter following completion of their proliferative potential, is the dominant phenotype in hybrids of Product Detail: normal and immortal cells. -

BMC Genomics Biomed Central
BMC Genomics BioMed Central Research article Open Access The functional modulation of epigenetic regulators by alternative splicing Sergio Lois1,2, Noemí Blanco1,2, Marian Martínez-Balbás*1,2 and Xavier de la Cruz*2,3 Address: 1Instituto de Biología Molecular de Barcelona. CID. Consejo Superior de Investigaciones Científicas (CSIC); 08028 Barcelona, Spain, 2Institut de Recerca Biomèdica-PCB; 08028 Barcelona, Spain and 3Institució Catalana de Recerca i Estudis Avançats (ICREA); Barcelona, Spain Email: Sergio Lois - [email protected]; Noemí Blanco - [email protected]; Marian Martínez-Balbás* - [email protected]; Xavier de la Cruz* - [email protected] * Corresponding authors Published: 25 July 2007 Received: 24 April 2007 Accepted: 25 July 2007 BMC Genomics 2007, 8:252 doi:10.1186/1471-2164-8-252 This article is available from: http://www.biomedcentral.com/1471-2164/8/252 © 2007 Lois et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Epigenetic regulators (histone acetyltransferases, methyltransferases, chromatin- remodelling enzymes, etc) play a fundamental role in the control of gene expression by modifying the local state of chromatin. However, due to their recent discovery, little is yet known about their own regulation. This paper addresses this point, focusing on alternative splicing regulation, a mechanism already known to play an important role in other protein families, e.g. transcription factors, membrane receptors, etc. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genome Analysis and Knowledge
Dahary et al. BMC Medical Genomics (2019) 12:200 https://doi.org/10.1186/s12920-019-0647-8 SOFTWARE Open Access Genome analysis and knowledge-driven variant interpretation with TGex Dvir Dahary1*, Yaron Golan1, Yaron Mazor1, Ofer Zelig1, Ruth Barshir2, Michal Twik2, Tsippi Iny Stein2, Guy Rosner3,4, Revital Kariv3,4, Fei Chen5, Qiang Zhang5, Yiping Shen5,6,7, Marilyn Safran2, Doron Lancet2* and Simon Fishilevich2* Abstract Background: The clinical genetics revolution ushers in great opportunities, accompanied by significant challenges. The fundamental mission in clinical genetics is to analyze genomes, and to identify the most relevant genetic variations underlying a patient’s phenotypes and symptoms. The adoption of Whole Genome Sequencing requires novel capacities for interpretation of non-coding variants. Results: We present TGex, the Translational Genomics expert, a novel genome variation analysis and interpretation platform, with remarkable exome analysis capacities and a pioneering approach of non-coding variants interpretation. TGex’s main strength is combining state-of-the-art variant filtering with knowledge-driven analysis made possible by VarElect, our highly effective gene-phenotype interpretation tool. VarElect leverages the widely used GeneCards knowledgebase, which integrates information from > 150 automatically-mined data sources. Access to such a comprehensive data compendium also facilitates TGex’s broad variant annotation, supporting evidence exploration, and decision making. TGex has an interactive, user-friendly, and easy adaptive interface, ACMG compliance, and an automated reporting system. Beyond comprehensive whole exome sequence capabilities, TGex encompasses innovative non-coding variants interpretation, towards the goal of maximal exploitation of whole genome sequence analyses in the clinical genetics practice. This is enabled by GeneCards’ recently developed GeneHancer, a novel integrative and fully annotated database of human enhancers and promoters. -

Understanding the Role of the Chromosome 15Q25.1 in COPD Through Epigenetics and Transcriptomics
European Journal of Human Genetics (2018) 26:709–722 https://doi.org/10.1038/s41431-017-0089-8 ARTICLE Understanding the role of the chromosome 15q25.1 in COPD through epigenetics and transcriptomics 1 1,2 1,3,4 5,6 5,6 Ivana Nedeljkovic ● Elena Carnero-Montoro ● Lies Lahousse ● Diana A. van der Plaat ● Kim de Jong ● 5,6 5,7 6 8 9 10 Judith M. Vonk ● Cleo C. van Diemen ● Alen Faiz ● Maarten van den Berge ● Ma’en Obeidat ● Yohan Bossé ● 11 1,12 12 1 David C. Nickle ● BIOS Consortium ● Andre G. Uitterlinden ● Joyce J. B. van Meurs ● Bruno C. H. Stricker ● 1,4,13 6,8 5,6 1 1 Guy G. Brusselle ● Dirkje S. Postma ● H. Marike Boezen ● Cornelia M. van Duijn ● Najaf Amin Received: 14 March 2017 / Revised: 6 November 2017 / Accepted: 19 December 2017 / Published online: 8 February 2018 © The Author(s) 2018. This article is published with open access Abstract Chronic obstructive pulmonary disease (COPD) is a major health burden in adults and cigarette smoking is considered the most important environmental risk factor of COPD. Chromosome 15q25.1 locus is associated with both COPD and smoking. Our study aims at understanding the mechanism underlying the association of chromosome 15q25.1 with COPD through epigenetic and transcriptional variation in a population-based setting. To assess if COPD-associated variants in 1234567890();,: 15q25.1 are methylation quantitative trait loci, epigenome-wide association analysis of four genetic variants, previously associated with COPD (P < 5 × 10−8) in the 15q25.1 locus (rs12914385:C>T-CHRNA3, rs8034191:T>C-HYKK, rs13180: C>T-IREB2 and rs8042238:C>T-IREB2), was performed in the Rotterdam study (n = 1489). -

Functional Mechanisms Underlying Pleiotropic Risk Alleles at the 19P13.1 Breast&Ndash;Ovarian Cancer Susceptibility Locus
ARTICLE Received 16 Jun 2015 | Accepted 20 Jul 2016 | Published 7 Sep 2016 DOI: 10.1038/ncomms12675 OPEN Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast–ovarian cancer susceptibility locus Kate Lawrenson et al.# A locus at 19p13 is associated with breast cancer (BC) and ovarian cancer (OC) risk. Here we analyse 438 SNPs in this region in 46,451 BC and 15,438 OC cases, 15,252 BRCA1 mutation carriers and 73,444 controls and identify 13 candidate causal SNPs associated with serous OC (P ¼ 9.2 Â 10 À 20), ER-negative BC (P ¼ 1.1 Â 10 À 13), BRCA1-associated BC (P ¼ 7.7 Â 10 À 16) and triple negative BC (P-diff ¼ 2 Â 10 À 5). Genotype-gene expression associations are identified for candidate target genes ANKLE1 (P ¼ 2 Â 10 À 3) and ABHD8 (Po2 Â 10 À 3). Chromosome conformation capture identifies interactions between four candidate SNPs and ABHD8, and luciferase assays indicate six risk alleles increased transactivation of the ADHD8 promoter. Targeted deletion of a region containing risk SNP rs56069439 in a putative enhancer induces ANKLE1 downregulation; and mRNA stability assays indicate functional effects for an ANKLE1 30-UTR SNP. Altogether, these data suggest that multiple SNPs at 19p13 regulate ABHD8 and perhaps ANKLE1 expression, and indicate common mechanisms underlying breast and ovarian cancer risk. Correspondence and requests for materials should be addressed to A.C.A. (email: [email protected]). #A full list of authors and their affiliations appears at the end of the paper. -

RNA Editing at Baseline and Following Endoplasmic Reticulum Stress
RNA Editing at Baseline and Following Endoplasmic Reticulum Stress By Allison Leigh Richards A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Human Genetics) in The University of Michigan 2015 Doctoral Committee: Professor Vivian G. Cheung, Chair Assistant Professor Santhi K. Ganesh Professor David Ginsburg Professor Daniel J. Klionsky Dedication To my father, mother, and Matt without whom I would never have made it ii Acknowledgements Thank you first and foremost to my dissertation mentor, Dr. Vivian Cheung. I have learned so much from you over the past several years including presentation skills such as never sighing and never saying “as you can see…” You have taught me how to think outside the box and how to create and explain my story to others. I would not be where I am today without your help and guidance. Thank you to the members of my dissertation committee (Drs. Santhi Ganesh, David Ginsburg and Daniel Klionsky) for all of your advice and support. I would also like to thank the entire Human Genetics Program, and especially JoAnn Sekiguchi and Karen Grahl, for welcoming me to the University of Michigan and making my transition so much easier. Thank you to Michael Boehnke and the Genome Science Training Program for supporting my work. A very special thank you to all of the members of the Cheung lab, past and present. Thank you to Xiaorong Wang for all of your help from the bench to advice on my career. Thank you to Zhengwei Zhu who has helped me immensely throughout my thesis even through my panic. -
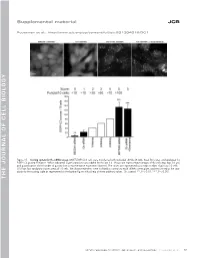
Thejournalofcellb Io Logy
Supplemental material JCB Puustinen et al., http://www.jcb.org/cgi/content/full/jcb.201304012/DC1 Figure S1. Scoring system for the siRNA screen. MCF7-EGFP-LC3 cells were transfected with indicated siRNAs (8 nM), fixed 56 h later, and analyzed for EGFP-LC3 puncta formation. When indicated, 5 µM siramesine was added for the last 3 h. Shown are representative images of the cells (top; bar, 10 µm) and quantification of the number of puncta from a representative experiment (bottom). The values are represented as a mean number of puncta/10 cells ± SDs from four randomly chosen areas of 10 cells. The shown treatments were included as controls to each siRNA screen plate, and they served as the stan- dards for the scoring scale as represented on the bottom figure with a help of three arbitrary values. Ctr, control. **, P < 0.01; ***, P < 0.001. THE JOURNAL OF CELL BIOLOGY CIP2A regulates mTORC1, cell growth, and autophagy • Puustinen et al. S1 Figure S2. A cartoon highlighting the positions of the candidate autophagy-regulating genes in the insulin receptor signaling network. The cartoon was prepared by Ingenuity Pathway Analysis. software. CP, carbohydrate phosphatase; ERK, extracellular signal–regulated kinase; FSH, follicle-stimulating hor- mone; EG, epidermal growth factor. S2 JCB Figure S3. The effect of siRNAs targeting the autophagy-regulating PP2 subunits on target gene expression, cell density, and mTORC1 pathway. (A) Total RNA was isolated from MCF7 cells treated with indicated siRNAs (20 nM) for 54 h and analyzed for target gene expression by qPCR using primers de- signed to specifically amplify the indicated PP2A-related mRNAs andACTB mRNA (internal control). -

Functional Mechanisms Underlying Pleiotropic Risk Alleles at the 19P13.1 Breast&Ndash;Ovarian Cancer Susceptibility Locus
UC Office of the President Recent Work Title Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast–ovarian cancer susceptibility locus Permalink https://escholarship.org/uc/item/1rx2f44n Author Lawrenson, Kate Publication Date 2016-09-07 DOI 10.1038/ncomms12675 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ARTICLE Received 16 Jun 2015 | Accepted 20 Jul 2016 | Published 7 Sep 2016 DOI: 10.1038/ncomms12675 OPEN Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast–ovarian cancer susceptibility locus Kate Lawrenson et al.# A locus at 19p13 is associated with breast cancer (BC) and ovarian cancer (OC) risk. Here we analyse 438 SNPs in this region in 46,451 BC and 15,438 OC cases, 15,252 BRCA1 mutation carriers and 73,444 controls and identify 13 candidate causal SNPs associated with serous OC (P ¼ 9.2 Â 10 À 20), ER-negative BC (P ¼ 1.1 Â 10 À 13), BRCA1-associated BC (P ¼ 7.7 Â 10 À 16) and triple negative BC (P-diff ¼ 2 Â 10 À 5). Genotype-gene expression associations are identified for candidate target genes ANKLE1 (P ¼ 2 Â 10 À 3) and ABHD8 (Po2 Â 10 À 3). Chromosome conformation capture identifies interactions between four candidate SNPs and ABHD8, and luciferase assays indicate six risk alleles increased transactivation of the ADHD8 promoter. Targeted deletion of a region containing risk SNP rs56069439 in a putative enhancer induces ANKLE1 downregulation; and mRNA stability assays indicate functional effects for an ANKLE1 30-UTR SNP. Altogether, these data suggest that multiple SNPs at 19p13 regulate ABHD8 and perhaps ANKLE1 expression, and indicate common mechanisms underlying breast and ovarian cancer risk. -

SPINT2 Suppresses Hippo Effector YAP and Limits Cellular Tolerance for Aneuploidy
SPINT2 Suppresses Hippo Effector YAP and Limits Cellular Tolerance for Aneuploidy The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Zhang, Huadi. 2017. SPINT2 Suppresses Hippo Effector YAP and Limits Cellular Tolerance for Aneuploidy. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:42061511 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA SPINT2 Suppresses Hippo Effector YAP and Limits Cellular Tolerance for Aneuploidy A dissertation presented by Huadi Zhang to The Division of Medical Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Biological and Biomedical Sciences Harvard University Cambridge, Massachusetts August 2017 © 2017 Huadi Zhang All rights reserved. Dissertation Advisor: Professor David Pellman Huadi Zhang SPINT2 Suppresses Hippo Effector YAP and Limits Cellular Tolerance for Aneuploidy Abstract Oncogenic transformation is often accompanied by chromosome instability, an increased rate of chromosome missegregation. The consequent gain or loss of chromosomes—termed aneuploidy—hinders the growth of most non-cancerous tissues, but is prevalent in tumors. During tumorigenesis, aneuploidy contributes to cellular heterogeneity and may promote downstream mutations, including chromosome rearrangements and oncogene amplification. Cellular mechanisms that safeguard against aneuploidy remain unclear. The Hippo pathway is a tumor-suppressor mechanism with essential roles in regulating tissue homeostasis. -

The Bio Revolution: Innovations Transforming and Our Societies, Economies, Lives
The Bio Revolution: Innovations transforming economies, societies, and our lives economies, societies, our and transforming Innovations Revolution: Bio The The Bio Revolution Innovations transforming economies, societies, and our lives May 2020 McKinsey Global Institute Since its founding in 1990, the McKinsey Global Institute (MGI) has sought to develop a deeper understanding of the evolving global economy. As the business and economics research arm of McKinsey & Company, MGI aims to help leaders in the commercial, public, and social sectors understand trends and forces shaping the global economy. MGI research combines the disciplines of economics and management, employing the analytical tools of economics with the insights of business leaders. Our “micro-to-macro” methodology examines microeconomic industry trends to better understand the broad macroeconomic forces affecting business strategy and public policy. MGI’s in-depth reports have covered more than 20 countries and 30 industries. Current research focuses on six themes: productivity and growth, natural resources, labor markets, the evolution of global financial markets, the economic impact of technology and innovation, and urbanization. Recent reports have assessed the digital economy, the impact of AI and automation on employment, physical climate risk, income inequal ity, the productivity puzzle, the economic benefits of tackling gender inequality, a new era of global competition, Chinese innovation, and digital and financial globalization. MGI is led by three McKinsey & Company senior partners: co-chairs James Manyika and Sven Smit, and director Jonathan Woetzel. Michael Chui, Susan Lund, Anu Madgavkar, Jan Mischke, Sree Ramaswamy, Jaana Remes, Jeongmin Seong, and Tilman Tacke are MGI partners, and Mekala Krishnan is an MGI senior fellow. -

Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling
International Journal of Molecular Sciences Review Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling Maria Carmela Annunziata 1, Melania Parisi 1, Gabriella Esposito 2 , Gabriella Fabbrocini 1, Rosario Ammendola 2 and Fabio Cattaneo 2,* 1 Department of Clinical Medicine and Surgery, School of Medicine, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (M.C.A.); [email protected] (M.P.); [email protected] (G.F.) 2 Department of Molecular Medicine and Medical Biotechnology, School of Medicine,, University of Naples Federico II, Via S. Pansini 5, 80131 Naples, Italy; [email protected] (G.E.); [email protected] (R.A.) * Correspondence: [email protected]; Fax: +39-081-7464-359 Received: 5 May 2020; Accepted: 25 May 2020; Published: 27 May 2020 Abstract: FPR1, FPR2, and FPR3 are members of Formyl Peptides Receptors (FPRs) family belonging to the GPCR superfamily. FPR2 is a low affinity receptor for formyl peptides and it is considered the most promiscuous member of this family. Intracellular signaling cascades triggered by FPRs include the activation of different protein kinases and phosphatase, as well as tyrosine kinase receptors transactivation. Protein kinases and phosphatases act coordinately and any impairment of their activation or regulation represents one of the most common causes of several human diseases. Several phospho-sites has been identified in protein kinases and phosphatases, whose role may be to expand the repertoire of molecular mechanisms of regulation or may be necessary for fine-tuning of switch properties. We previously performed a phospho-proteomic analysis in FPR2-stimulated cells that revealed, among other things, not yet identified phospho-sites on six protein kinases and one protein phosphatase.