Network Complexity of Foodwebs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Guiding Organic Management in a Service-Oriented Real-Time Middleware Architecture
Guiding Organic Management in a Service-Oriented Real-Time Middleware Architecture Manuel Nickschas and Uwe Brinkschulte Institute for Computer Science University of Frankfurt, Germany {nickschas, brinks}@es.cs.uni-frankfurt.de Abstract. To cope with the ever increasing complexity of today's com- puting systems, the concepts of organic and autonomic computing have been devised. Organic or autonomic systems are characterized by so- called self-X properties such as self-conguration and self-optimization. This approach is particularly interesting in the domain of distributed, embedded, real-time systems. We have already proposed a service-ori- ented middleware architecture for such systems that uses multi-agent principles for implementing the organic management. However, organic management needs some guidance in order to take dependencies between services into account as well as the current hardware conguration and other application-specic knowledge. It is important to allow the appli- cation developer or system designer to specify such information without having to modify the middleware. In this paper, we propose a generic mechanism based on capabilities that allows describing such dependen- cies and domain knowledge, which can be combined with an agent-based approach to organic management in order to realize self-X properties. We also describe how to make use of this approach for integrating the middleware's organic management with node-local organic management. 1 Introduction Distributed, embedded systems are rapidly advancing in all areas of our lives, forming increasingly complex networks that are increasingly hard to handle for both system developers and maintainers. In order to cope with this explosion of complexity, also commonly referred to as the Software Crisis [1], the concepts of Autonomic [2,3] and Organic [4,5,6] Computing have been devised. -

Organic Computing Doctoral Dissertation Colloquium 2018 B
13 13 S. Tomforde | B. Sick (Eds.) Organic Computing Doctoral Dissertation Colloquium 2018 Organic Computing B. Sick (Eds.) B. | S. Tomforde S. Tomforde ISBN 978-3-7376-0696-7 9 783737 606967 V`:%$V$VGVJ 0QJ `Q`8 `8 V`J.:`R 1H@5 J10V`1 ? :VC Organic Computing Doctoral Dissertation Colloquium 2018 S. Tomforde, B. Sick (Editors) !! ! "#$%&'&$'$(&)(#(&$* + "#$%&'&$'$(&)(#$&,*&+ - .! ! /)!/#0// 12#$%'$'$()(#$, 23 & ! )))0&,)(#$' '0)/#5 6 7851 999!& ! kassel university press 7 6 Preface The Organic Computing Doctoral Dissertation Colloquium (OC-DDC) series is part of the Organic Computing initiative [9, 10] which is steered by a Special Interest Group of the German Society of Computer Science (Gesellschaft fur¨ Informatik e.V.). It provides an environment for PhD students who have their research focus within the broader Organic Computing (OC) community to discuss their PhD project and current trends in the corresponding research areas. This includes, for instance, research in the context of Autonomic Computing [7], ProActive Computing [11], Complex Adaptive Systems [8], Self-Organisation [2], and related topics. The main goal of the colloquium is to foster excellence in OC-related research by providing feedback and advice that are particularly relevant to the students’ studies and ca- reer development. Thereby, participants are involved in all stages of the colloquium organisation and the review process to gain experience. The OC-DDC 2018 took place in -

Teaching and Learning with Digital Evolution: Factors Influencing Implementation and Student Outcomes
TEACHING AND LEARNING WITH DIGITAL EVOLUTION: FACTORS INFLUENCING IMPLEMENTATION AND STUDENT OUTCOMES By Amy M Lark A DISSERTATION Submitted to Michigan State University in partial fulfillment of the requirements for the degree of Curriculum, Instruction, and Teacher Education – Doctor of Philosophy 2014 ABSTRACT TEACHING AND LEARNING WITH DIGITAL EVOLUTION: FACTORS INFLUENCING IMPLEMENTATION AND STUDENT OUTCOMES By Amy M Lark Science literacy for all Americans has been the rallying cry of science education in the United States for decades. Regardless, Americans continue to fall short when it comes to keeping pace with other developed nations on international science education assessments. To combat this problem, recent national reforms have reinvigorated the discussion of what and how we should teach science, advocating for the integration of disciplinary core ideas, crosscutting concepts, and science practices. In the biological sciences, teaching the core idea of evolution in ways consistent with reforms is fraught with challenges. Not only is it difficult to observe biological evolution in action, it is nearly impossible to engage students in authentic science practices in the context of evolution. One way to overcome these challenges is through the use of evolving populations of digital organisms. Avida-ED is digital evolution software for education that allows for the integration of science practice and content related to evolution. The purpose of this study was to investigate the effects of Avida-ED on teaching and learning evolution and the nature of science. To accomplish this I conducted a nationwide, multiple-case study, documenting how instructors at various institutions were using Avida-ED in their classrooms, factors influencing implementation decisions, and effects on student outcomes. -

Evolving Quorum Sensing in Digital Organisms
Evolving Quorum Sensing in Digital Organisms Benjamin E. Beckmann and Phillip K. McKinley Department of Computer Science and Engineering 3115 Engineering Building Michigan State University East Lansing, Michigan 48824 {beckma24,mckinley}@cse.msu.edu ABSTRACT 1. INTRODUCTION For centuries it was thought that bacteria live asocial lives. The human body is made up of 1013 human cells. Yet, However, recent discoveries show many species of bacteria this number is an order of magnitude smaller than the num- communicate in order to perform tasks previously thought ber a bacterial cells living within the gastrointestinal tract to be limited to multicellular organisms. Central to this ca- of a single adult [4]. Although it was previously assumed pability is quorum sensing, whereby organisms detect cell that bacteria and other microorganisms rarely interact [24], density and use this information to trigger group behav- in 1979 Nealson and Hastings found evidence that bacterial iors. Quorum sensing is used by bacteria in the formation communities of Vibrio fischeri and Vibrio harveyi were able of biofilms, secretion of digestive enzymes and, in the case to perform a coordinated behavioral change, namely emit- of pathogenic bacteria, release of toxins or other virulence ting light, when the cell density rose above a certain thresh- factors. Indeed, methods to disrupt quorum sensing are cur- old [20]. This type of density-based behavioral change is rently being investigated as possible treatments for numer- called quorum sensing [27]. In quorum sensing (QS), con- ous diseases, including cystic fibrosis, epidemic cholera, and tinual secretion and detection of chemicals provide a way methicillin-resistant Staphylococcus aureus. -

Digital Irreducible Complexity: a Survey of Irreducible Complexity in Computer Simulations
Critical Focus Digital Irreducible Complexity: A Survey of Irreducible Complexity in Computer Simulations Winston Ewert* Biologic Institute, Redmond, Washington, USA Abstract Irreducible complexity is a concept developed by Michael Behe to describe certain biological systems. Behe claims that irreducible complexity poses a challenge to Darwinian evolution. Irreducibly complex systems, he argues, are highly unlikely to evolve because they have no direct series of selectable intermediates. Various computer models have been published that attempt to demonstrate the evolution of irreducibly complex systems and thus falsify this claim. How- ever, closer inspection of these models shows that they fail to meet the definition of irreducible complexity in a num- ber of ways. In this paper we demonstrate how these models fail. In addition, we present another designed digital sys- tem that does exhibit designed irreducible complexity, but that has not been shown to be able to evolve. Taken together, these examples indicate that Behe’s concept of irreducible complexity has not been falsified by computer models. Cite as: Ewert W (2014) Digital irreducible complexity: A survey of irreducible complexity in computer simulations. BIO-Complexity 2014 (1):1–10. doi:10.5048/BIO-C.2014.1. Editor: Ola Hössjer Received: November 16, 2013; Accepted: February 18, 2014; Published: April 5, 2014 Copyright: © 2014 Ewert. This open-access article is published under the terms of the Creative Commons Attribution License, which permits free distribution and reuse in derivative works provided the original author(s) and source are credited. Notes: A Critique of this paper, when available, will be assigned doi:10.5048/BIO-C.2014.1.c. -

Applying Evolutionary Computation to Mitigate Uncertainty in Dynamically-Adaptive, High-Assurance Middleware
J Internet Serv Appl (2012) 3:51–58 DOI 10.1007/s13174-011-0049-4 SI: FOME - THE FUTURE OF MIDDLEWARE Applying evolutionary computation to mitigate uncertainty in dynamically-adaptive, high-assurance middleware Philip K. McKinley · Betty H.C. Cheng · Andres J. Ramirez · Adam C. Jensen Received: 31 October 2011 / Accepted: 12 November 2011 / Published online: 3 December 2011 © The Brazilian Computer Society 2011 Abstract In this paper, we explore the integration of evolu- systems need to perform complex, distributed tasks despite tionary computation into the development and run-time sup- lossy wireless communication, limited resources, and uncer- port of dynamically-adaptable, high-assurance middleware. tainty in sensing the environment. However, even systems The open-ended nature of the evolutionary process has been that operate entirely within cyberspace need to account for shown to discover novel solutions to complex engineering changing network and load conditions, component failures, problems. In the case of high-assurance adaptive software, and exposure to a wide variety of cyber attacks. however, this search capability must be coupled with rigor- A dynamically adaptive system (DAS) monitors itself and ous development tools and run-time support to ensure that its execution environment at run time. This monitoring en- the resulting systems behave in accordance with require- ables a DAS not only to detect conditions warranting a re- ments. Early investigations are reviewed, and several chal- configuration, but also to determine when and how to safely lenging problems and possible research directions are dis- reconfigure itself in order to deliver acceptable behavior be- cussed. fore, during, and after adaptation [2]. -

Editorial on Digital Organism
Editorial Journal of Computer Science & Volume 13:6, 2020 DOI: 10.37421/jcsb.2020.13.325 Systems Biology ISSN: 0974-7230 Open Access Editorial on Digital Organism Chinthala Mounica* Department of Computer Science, Osmania University, India a growing number of evolutionary biologists. Evolutionary biologist Richard Editorial Note Lenski of Michigan State University has used Avida extensively in his work. Lenski, Adami, and their colleagues have published in journals such as Nature An advanced creature is a self-duplicating PC program that changes and and the Proceedings of the National Academy of Sciences (USA). Digital develops. Advanced creatures are utilized as an apparatus to contemplate the organisms can be traced back to the game Darwin, developed in 1961 at elements of Darwinian development, and to test or check explicit speculations Bell Labs, in which computer programs had to compete with each other by or numerical models of development. The investigation of computerized trying to stop others from executing. A similar implementation that followed creatures is firmly identified with the region of counterfeit life. this was the game Core War. In Core War, it turned out that one of the winning Digital organisms can be traced back to the game Darwin, developed in strategies was to replicate as fast as possible, which deprived the opponent of 1961 at Bell Labs, in which computer programs had to compete with each other all computational resources. Programs in the Core War game were also able to by trying to stop others from executing. A similar implementation that followed mutate themselves and each other by overwriting instructions in the simulated this was the game Core War. -

Evolution of Biological Complexity
Evolution of biological complexity Christoph Adami*†, Charles Ofria‡§, and Travis C. Collier¶ *Kellogg Radiation Laboratory 106-38 and ‡Beckman Institute 139-74, California Institute of Technology, Pasadena, CA 91125; and ¶Division of Organismic Biology, Ecology, and Evolution, University of California, Los Angeles, CA 90095 Edited by James F. Crow, University of Wisconsin, Madison, WI, and approved February 15, 2000 (received for review December 22, 1999) To make a case for or against a trend in the evolution of complexity distinguish distinct evolutionary pressures acting on the genome in biological evolution, complexity needs to be both rigorously and analyze them in a mathematical framework. defined and measurable. A recent information-theoretic (but intu- If an organism’s complexity is a reflection of the physical itively evident) definition identifies genomic complexity with the complexity of its genome (as we assume here), the latter is of amount of information a sequence stores about its environment. prime importance in evolutionary theory. Physical complexity, We investigate the evolution of genomic complexity in populations roughly speaking, reflects the number of base pairs in a sequence of digital organisms and monitor in detail the evolutionary tran- that are functional. As is well known, equating genomic com- sitions that increase complexity. We show that, because natural plexity with genome length in base pairs gives rise to a conun- selection forces genomes to behave as a natural ‘‘Maxwell De- drum (known as the C-value paradox) because large variations mon,’’ within a fixed environment, genomic complexity is forced to in genomic complexity (in particular in eukaryotes) seem to bear increase. -

Analysis of Evolutionary Algorithms
Why We Do Not Evolve Software? Analysis of Evolutionary Algorithms Roman V. Yampolskiy Computer Engineering and Computer Science University of Louisville [email protected] Abstract In this paper, we review the state-of-the-art results in evolutionary computation and observe that we don’t evolve non-trivial software from scratch and with no human intervention. A number of possible explanations are considered, but we conclude that computational complexity of the problem prevents it from being solved as currently attempted. A detailed analysis of necessary and available computational resources is provided to support our findings. Keywords: Darwinian Algorithm, Genetic Algorithm, Genetic Programming, Optimization. “We point out a curious philosophical implication of the algorithmic perspective: if the origin of life is identified with the transition from trivial to non-trivial information processing – e.g. from something akin to a Turing machine capable of a single (or limited set of) computation(s) to a universal Turing machine capable of constructing any computable object (within a universality class) – then a precise point of transition from non-life to life may actually be undecidable in the logical sense. This would likely have very important philosophical implications, particularly in our interpretation of life as a predictable outcome of physical law.” [1]. 1. Introduction On April 1, 2016 Dr. Yampolskiy posted the following to his social media accounts: “Google just announced major layoffs of programmers. Future software development and updates will be done mostly via recursive self-improvement by evolving deep neural networks”. The joke got a number of “likes” but also, interestingly, a few requests from journalists for interviews on this “developing story”. -

Evolution of Stable Ecosystems in Populations of Digital Organisms
in Artificial Life VIII, Standish, Abbass, Bedau (eds)(MIT Press) 2002. pp 227{232 1 Evolution of Stable Ecosystems in Populations of Digital Organisms Tim F. Cooper and Charles Ofria Center for Microbial Ecology Michigan State University, East Lansing, MI 48824 Abstract continued use will diminish and organisms able to use un- derutilized resources will be favored. Over time density- Competition for resources has long been believed to be dependent selection created by this process can lead to fundamental to the evolution of diversity. However, the difficulty of working with natural ecosystems has meant adaptive radiation and even speciation as organisms di- that this theory has rarely been tested. Here, we use the verge to into distinct lineages (Schluter 2001). Avida experimental platform to demonstrate the evolu- In this study, we use digital organisms (self-replicating tion of ecosystems composed of digital organisms. We and evolving computer programs) to examine the di- show that stable ecosystems are formed during evolu- versity arising in populations during adaptation to an tion in an environment where organisms must compete for limiting resources. Stable coexistence was not ob- environment containing nine distinct resources. (We served in environments where resource levels were not use the term population to refer collectively to all or- limiting, suggesting that competition for resources was ganisms in the environment.) We compare the result responsible for coexistence. To test this, we restarted of adaptation when resources are in infinite supply to populations evolved in the resource-limited environment when resources can be depleted. Because there are but increased resource levels to be non-limiting. -

IJACSIT Volume 03
Contents Editorial Board V I-I Holistic Electronic Government Services Integration Model: from Theory to Practice Tadas Limba, Gintarė Gulevičiūtė 1-31 Detecting Suspicion Information on the Web Using Crime Data Mining Techniques Javad Hosseinkhani, Mohammad Koochakzaei, Solmaz Keikhaee, Yahya Hamedi Amin 32-41 Develop a New Method for People Identification Using Hand Appearance Mahdi Nasrollah Barati, Seyed Yaser Bozorgi Rad 42-49 Model and Solve the Bi-Criteria Multi Source Flexible Multistage Logistics Network Seyed Yaser Bozorgi Rad, Mohammad Ishak Desa, Sara Delfan Azari 50-69 Impact of Strategic Management Element in Enhancing Firm’s Sustainable Competitive Advantage. An Empirical Study of Nigeria’s Manufacturing Sector Yahaya Sani, Abdel-Hafiez Ali Hassaballah 70-82 A Co-modal Transport Information System in a Distributed Environment Zhanjun Wang, Khaled Mesghouni, Slim Hammadi 83-99 Online Brand Experience Creation Process Model: Theoretical Insights Tadas Limba, Mindaugas Kiskis, Virginija Jurkute 100-118 Color Image Segmentation Using a Modified Fuzzy C-means Method and Data Fusion Techniques Rafika Harrabi, Ezzedine Ben Braiek 119-134 Model of Brand Building and Enhancement by Electronic Marketing Tools: Practical Implication Tadas Limba, Gintarė Gulevičiūtė, Virginija Jurkutė 135-155 A Constraint Programming Approach for Scheduling in a Multi-Project Environment Marcin Relich 156-171 Anti-Crisis Management Tools for Capitalist Economy Alexander A. Antonov 172-190 E-Business Qualitative Criteria Application Model: Perspectives of Practical Implementation Tadas Limba, Gintarė Gulevičiūtė 191-213 A UML Profile for use cases Multi-interpretation Mira Abboud, Hala Naja, Mohamad Dbouk, Bilal El Haj Ali 214-226 A Grid-enabled Application for the Simulation of Plant Tissue Culture Experiments Florence I. -
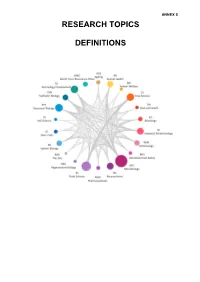
Research Topic Definitions
ANNEX 5 RESEARCH TOPICS DEFINITIONS Table of Contents Ageing (AGE) ..........................................................................................................................3 Animal Health (AH) ................................................................................................................5 Animal Welfare (AW) .............................................................................................................6 EG (EG) ..................................................................................................................................7 Crop Science (CS) .................................................................................................................8 Diet and Health (DH) ..............................................................................................................9 Immunology (IMM) ...............................................................................................................10 Industrial Biotechnology (IB) .............................................................................................. 11 Microbial Food Safety (MFS) .............................................................................................. 12 Microbiology (MIC) ..............................................................................................................13 Neuroscience and Behaviour (NS) .....................................................................................14 Pharmaceuticals (PHM) .......................................................................................................14