SURYA-DISSERTATION-2018.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Band 49 • Heft 2 • Mai 2011
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Hochschulschriftenserver - Universität Frankfurt am Main Band 49 • Heft 2 • Mai 2011 Deutsche Ornithologen-Gesellschaft e.V. Institut für Vogelforschung Vogelwarte Hiddensee Max-Planck-Institut für Ornithologie „Vogelwarte Helgoland“ und Vogelwarte Radolfzell Beringungszentrale Hiddensee Die „Vogelwarte“ ist offen für wissenschaftliche Beiträge und Mitteilungen aus allen Bereichen der Orni- tho logie, einschließlich Avifaunistik und Beringungs wesen. Zusätzlich zu Originalarbeiten werden Kurz- fassungen von Dissertationen, Master- und Diplomarbeiten aus dem Be reich der Vogelkunde, Nach richten und Terminhinweise, Meldungen aus den Berin gungszentralen und Medienrezensionen publiziert. Daneben ist die „Vogelwarte“ offizielles Organ der Deutschen Ornithologen-Gesellschaft und veröffentlicht alle entsprechenden Berichte und Mitteilungen ihrer Gesellschaft. Herausgeber: Die Zeitschrift wird gemein sam herausgegeben von der Deutschen Ornithologen-Gesellschaft, dem Institut für Vogelforschung „Vogelwarte Helgoland“, der Vogelwarte Radolfzell am Max-Planck-Institut für Ornithologie, der Vogelwarte Hiddensee und der Beringungszentrale Hiddensee. Die Schriftleitung liegt bei einem Team von vier Schriftleitern, die von den Herausgebern benannt werden. Die „Vogelwarte“ ist die Fortsetzung der Zeitschriften „Der Vogelzug“ (1930 – 1943) und „Die Vogelwarte“ (1948 – 2004). Redaktion / Schriftleitung: DO-G-Geschäftsstelle: Manuskripteingang: Dr. Wolfgang Fiedler, Vogelwarte Radolf- Karl Falk, c/o Institut für Vogelfor- zell am Max-Planck-Institut für Ornithologie, Schlossallee 2, schung, An der Vogelwarte 21, 26386 D-78315 Radolfzell (Tel. 07732/1501-60, Fax. 07732/1501-69, Wilhelmshaven (Tel. 0176/78114479, [email protected]) Fax. 04421/9689-55, Dr. Ommo Hüppop, Institut für Vogelforschung „Vogelwarte Hel- [email protected] http://www.do-g.de) goland“, Inselstation Helgoland, Postfach 1220, D-27494 Helgo- Alle Mitteilungen und Wünsche, welche die Deutsche Ornitho- land (Tel. -

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

Sichuan, China
Tropical Birding: Sichuan (China). Custom Tour Report A Tropical Birding custom tour SICHUAN, CHINA : (Including the Southern Shans Pre-tour Extension) WHITE-THROATED TIT One of 5 endemic tits recorded on the tour. 21 May – 12 June, 2010 Tour Leader: Sam Woods All photos were taken by Sam Woods/Tropical Birding on this tour, except one photo. www.tropicalbirding.com [email protected] 1-409-515-0514 Tropical Birding: Sichuan (China). Custom Trip Report The Central Chinese province of Sichuan provided some notable challenges this year: still recovering from the catastrophic “Wenchuan 5.12” earthquake of 2008, the area is undergoing massive reconstruction. All very positive for the future of this scenically extraordinary Chinese region, but often a headache for tour arrangements, due to last minute traffic controls leading us to regularly rethink our itinerary in the Wolong area in particular, that was not far from the epicenter of that massive quake. Even in areas seemingly unaffected by the quake, huge road construction projects created similar challenges to achieving our original planned itinerary. However, in spite of regular shuffling and rethinking, the itinerary went ahead pretty much as planned with ALL sites visited. Other challenges came this year in the form of heavy regular rains that plagued us at Wawu Shan and low cloud that limited visibility during our time around the breathtaking Balang Mountain in the Wolong region. With some careful trickery, sneaking our way through week-long road blocks under cover of darkness, birding through thick and thin (mist, cloud and rains) we fought against all such challenges and came out on top. -

Andhra Pradesh
PROFILES OF SELECTED NATIONAL PARKS AND SANCTUARIES OF INDIA JULY 2002 EDITED BY SHEKHAR SINGH ARPAN SHARMA INDIAN INSTITUTE OF PUBLIC ADMINISTRATION NEW DELHI CONTENTS STATE NAME OF THE PA ANDAMAN AND NICOBAR CAMPBELL BAY NATIONAL PARK ISLANDS GALATHEA NATIONAL PARK MOUNT HARRIET NATIONAL PARK NORTH BUTTON ISLAND NATIONAL PARK MIDDLE BUTTON ISLAND NATIONAL PARK SOUTH BUTTON ISLAND NATIONAL PARK RANI JHANSI MARINE NATIONAL PARK WANDOOR MARINE NATIONAL PARK CUTHBERT BAY WILDLIFE SANCTUARY GALATHEA BAY WILDLIFE SANCTUARY INGLIS OR EAST ISLAND SANCTUARY INTERVIEW ISLAND SANCTUARY LOHABARRACK OR SALTWATER CROCODILE SANCTUARY ANDHRA PRADESH ETURUNAGARAM SANCTUARY KAWAL WILDLIFE SANCTUARY KINNERSANI SANCTUARY NAGARJUNASAGAR-SRISAILAM TIGER RESERVE PAKHAL SANCTUARY PAPIKONDA SANCTUARY PRANHITA WILDLIFE SANCTUARY ASSAM MANAS NATIONAL PARK GUJARAT BANSDA NATIONAL PARK PURNA WILDLIFE SANCTUARY HARYANA NAHAR SANCTUARY KALESAR SANCTUARY CHHICHHILA LAKE SANCTUARY ABUBSHEHAR SANCTUARY BIR BARA VAN JIND SANCTUARY BIR SHIKARGAH SANCTUARY HIMACHAL PRADESH PONG LAKE SANCTUARY RUPI BHABA SANCTUARY SANGLA SANCTUARY KERALA SILENT VALLEY NATIONAL PARK ARALAM SANCTUARY CHIMMONY SANCTUARY PARAMBIKULAM SANCTUARY PEECHI VAZHANI SANCTUARY THATTEKAD BIRD SANCTUARY WAYANAD WILDLIFE SANCTUARY MEGHALAYA BALPAKARAM NATIONAL PARK SIJU WILDLIFE SANCTUARY NOKREK NATIONAL PARK NONGKHYLLEM WILDLIFE SANCTUARY MIZORAM MURLEN NATIONAL PARK PHAWNGPUI (BLUE MOUNTAIN) NATIONAL 2 PARK DAMPA WILDLIFE SANCTUARY KHAWNGLUNG WILDLIFE SANCTUARY LENGTENG WILDLIFE SANCTUARY NGENGPUI WILDLIFE -

Sri Lanka: January 2015
Tropical Birding Trip Report Sri Lanka: January 2015 A Tropical Birding CUSTOM tour SRI LANKA: Ceylon Sojourn 9th- 23rd January 2015 Tour Leaders: Sam Woods & Chaminda Dilruk SRI LANKA JUNGLEFOWL is Sri Lanka’s colorful national bird, which was ranked among the top five birds of the tour by the group. All photos in this report were taken by Sam Woods. 1 www.tropicalbirding.com +1-409-515-0514 [email protected] Page Tropical Birding Trip Report Sri Lanka: January 2015 INTRODUCTION In many ways Sri Lanka covers it all; for the serious birder, even those with experience from elsewhere in the Indian subcontinent, it offers up a healthy batch of at least 32 endemic bird species (this list continues to grow, though, so could increase further yet); for those without any previous experience of the subcontinent it offers these but, being an island of limited diversity, not the overwhelming numbers of birds, which can be intimidating for the first timer; and for those with a natural history slant that extends beyond the avian, there is plentiful other wildlife besides, to keep all happy, such as endemic monkeys, strange reptiles only found on this teardrop-shaped island, and a bounty of butterflies, which feature day-in, day-out. It should also be made clear that while it appears like a chunk of India which has dropped of the main subcontinent, to frame it, as merely an extension of India, would be a grave injustice, as Sri Lanka feels, looks, and even tastes very different. There are some cultural quirks that make India itself, sometimes challenging to visit for the westerner. -
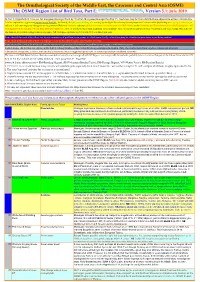
ORL 5.1 Hypothetical Spp Final Draft01a.Xlsx
The Ornithological Society of the Middle East, the Caucasus and Central Asia (OSME) The OSME Region List of Bird Taxa, Part E: , Version 5.1: July 2019 In Part E, Hypothetical Taxa, we list non-passerines (prefixed by 'N') first, then passerines (prefixed by 'P'). Such taxa may be from distributions adjacent to or have extended to A fuller explanation is given in Explanation of the ORL, but briefly, Bright green shading of a row (eg Syrian Ostrich) indicates former presence of a taxon in the OSME Region. Light gold shading in column A indicates sequence change from the previous ORL issue. Red font indicates added information since the previous ORL version or the Conservation Threat Status (Critically Endangered = CE, Endangered = E, Vulnerable = V and Data Deficient = DD only). Not all synonyms have been examined. Serial numbers (SN) are merely an administrative convenience and may change. Please do not cite them in any formal correspondence or papers. NB: Compass cardinals (eg N = north, SE = southeast) are used. Rows shaded thus and with yellow text denote summaries of problem taxon groups in which some closely-related taxa may be of indeterminate status or are being studied. Rows shaded thus and with yellow text indicate recent or data-driven major conservation concerns. Rows shaded thus and with white text contain additional explanatory information on problem taxon groups as and when necessary. English names shaded thus are species on BirdLife Tracking Database, http://seabirdtracking.org/mapper/index.php. Only a few individuals from very few colonies are involved. A broad dark orange line, as below, indicates the last taxon in a new or suggested species split, or where sspp are best considered separately. -

West & East Sikkim November
West & East Sikkim_November 2018 Date: 3rd November 2018 to 9th November 2018 Habitat: Broad-leaf / Temperate Coniferous. Montane Forest. Conifers / Alpine. Dwarf Junipers and Dwarf Rhododendron Steppe Grassland. Riverine / Streams / Dam. Alluvial. Human Habitat. Temperature Range: -2°C ~ 25°C Places: Melli-Jorethang-Legship-Tashiding-Yuksom-KNP in West Sikkim District Rongpo-Rongli-Lingtham-Phadamchen-Zuluk-Kupup-Sherathang in East Sikkim District Bird Checklist (As per Birds of Indian Subcontinent field guide by Richard Grimmett, Carol Inskipp, Tim Inskipp): 1. Rufous-throated Partridge - Heard multiple times on day 6 (on 8th November) while coming back to home stay after morning birding session. Call was coming from nearby hillock from inaccessible location. 2. Himalayan Monal - Multiple sightings. First sight was on day 5 (on 7th November), a female crossing a ditch. No photographs taken. Later in the afternoon near Lungthu a flock of five females were seen. Successfully photographed. On 8th November the morning session was dedicated for Monal male. Sighted the male for more than 30 minutes; with the male five other individuals (female+juvenile) was seen. Entire flock was moving high up. 3. Bar-headed Geese - Two birds sighted at Elephant Lake (Bedang Tso) at the northern extreme of the sanctuary. 4. Ruddy Shelduck - 32 Individuals seen along with Bar-headed Geese foraging on algae lake, adjacent to Bedang Tso. 5. Little Egret - Saw around 10 of them while crossing eastern range of Mahananda on road adjacent to Teesta riverbed. 6. Indian Cormorant - On Teesta - Sevok rail gate area, some perched on rock / boulders. 7. Great Cormorant - Saw on the way to Melli on day 1 (3rd Nov) just before entering Sikkim check post. -

SICHUAN (Including Northern Yunnan)
Temminck’s Tragopan (all photos by Dave Farrow unless indicated otherwise) SICHUAN (Including Northern Yunnan) 16/19 MAY – 7 JUNE 2018 LEADER: DAVE FARROW The Birdquest tour to Sichuan this year was a great success, with a slightly altered itinerary to usual due to the closure of Jiuzhaigou, and we enjoyed a very smooth and enjoyable trip around the spectacular and endemic-rich mountain and plateau landscapes of this striking province. Gamebirds featured strongly with 14 species seen, the highlights of them including a male Temminck’s Tragopan grazing in the gloom, Chinese Monal trotting across high pastures, White Eared and Blue Eared Pheasants, Lady Amherst’s and Golden Pheasants, Chinese Grouse and Tibetan Partridge. Next were the Parrotbills, with Three-toed, Great and Golden, Grey-hooded and Fulvous charming us, Laughingthrushes included Red-winged, Buffy, Barred, Snowy-cheeked and Plain, we saw more Leaf Warblers than we knew what to do with, and marvelled at the gorgeous colours of Sharpe’s, Pink-rumped, Vinaceous, Three-banded and Red-fronted Rosefinches, the exciting Przevalski’s Finch, the red pulse of Firethroats plus the unreal blue of Grandala. Our bird of the trip? Well, there was that Red Panda that we watched for ages! 1 BirdQuest Tour Report: Sichuan Including Northern Yunnan 2018 www.birdquest-tours.com Our tour began with a short extension in Yunnan, based in Lijiang city, with the purpose of finding some of the local specialities including the rare White-speckled Laughingthrush, which survives here in small numbers. Once our small group had arrived in the bustling city of Lijiang we began our birding in an area of hills that had clearly been totally cleared of forest in the fairly recent past, with a few trees standing above the hillsides of scrub. -

The Generic Taxonomy of Parrotbills (Aves, Timaliidae)
FORKTAIL 25 (2009): 137–141 The generic taxonomy of parrotbills (Aves, Timaliidae) JOHN PENHALLURICK and CRAIG ROBSON The parrotbills are typically considered to contain just three genera: Conostoma, Paradoxornis and Panurus. Discounting Panurus from consideration (it has recently been shown to have a distant relationship to the babblers), we maintain a single species in Conostoma, C. aemodium, and assign the species currently lumped into Paradoxornis among seven genera that fall into two groups based in part on size: the first group (which also includes Conostoma) consists of Hemirhynchus (for paradoxus and unicolor); Psittiparus (for gularis, margaritae, ruficeps and bakeri) and Paradoxornis (for flavirostris, guttaticollis and heudei); the second comprises Chleuasicus (for atrosuperciliaris), a new genus Sinosuthora (for brunnea, webbiana, alphonsiana, conspicillata, zappeyi and przewalskii), Neosuthora (for davidiana) and Suthora (for fulvifrons, verreauxi, nipalensis, humii, poliotis, ripponi and beaulieui). INTRODUCTION to reflect this distant relationship. We will go through the genera we propose, giving the full citation for the generic Earlier accounts of the parrotbills, such as Sharpe (1883), name, plus synonyms, and listing the species we assign to Hartert (1907), Hartert and Steinbacher (1932–38), and each genus, and its subspecies, with detailed distribution Baker (1930), treated them in multiple genera, but in provided for both monotypic species and subspecies. recent works (Deignan 1964, Dickinson 2003, Robson 2007) the great majority have been placed in Paradoxornis. This arrangement goes back to Delacour (1946), who THE GENERA AND SPECIES assigned all taxa except Great Parrotbill Conostoma aemodium and Bearded Reedling Panurus biarmicus to Conostoma Hodgson, 1842 Paradoxornis. His explanation for this radical move was Conostoma Hodgson, 1842 [‘1841’], Journal of the Asiatic brief, and roughly translates as follows: Society of Bengal 10: 856. -

Bird Checklists of the World Country Or Region: Myanmar
Avibase Page 1of 30 Col Location Date Start time Duration Distance Avibase - Bird Checklists of the World 1 Country or region: Myanmar 2 Number of species: 1088 3 Number of endemics: 5 4 Number of breeding endemics: 0 5 Number of introduced species: 1 6 7 8 9 10 Recommended citation: Lepage, D. 2021. Checklist of the birds of Myanmar. Avibase, the world bird database. Retrieved from .https://avibase.bsc-eoc.org/checklist.jsp?lang=EN®ion=mm [23/09/2021]. Make your observations count! Submit your data to ebird. -

On the Base of Molecular Data, a Great Difference Between Sedentary
ISSN 2347-6893 On the base of molecular data, a great difference between sedentary subspecies of Prinia subflava Gmelin, 1789 suggests resurrecting name Prinia mutatrix Meise, 1936 Billy Nguembock1,2,*, Mahamat Sali1, Azang Esther Diane Olivia1, Guehoada Yollande1 1Laboratory of Zoology, Ornithology Unit, Department of Animal Biology and Physiology, University of Yaounde I, P.O. Box 812 Yaounde, Cameroon; 2Department of Biological Sciences, University of Montreal, C.P. 6128, Center-town, Montreal, Quebec, H3C 3J7, Canada. ABSTRACT Prinia subflava is a sedentary member of the “open warblers cisticolid” clade and in our first papers, we noted a strong divergence between Prinia subflava subflava and Prinia subflava mutatrix. To confirm this divergence and leaning partially on our first obtained results, we investigated the genetic variation of the individuals of P. s. subflava (West Africa) and P. s. mutatrix (South East Africa). For the genetic variation, we used three mitochondrial genes (ATPase6, ND2 and ND3) to calculate their genetic distances within the cisticolid ingroup and to explore their mutational differentiation. With our ATPase6, ND2 and ND3, a genetic distance of 5.76%, 5.15% and 5.13% was estimated respectively between individuals of P. s. subflava (Cameroon) and P. s. mutatrix (Malawi) whereas it was, for the protein-coding gene ND2, only of 1.81% between the specimens of P. s. subflava caught in parts of West Africa (Gambia and Cameroon). For the mutational differentiation, a total of 113 different molecular characters were observed on the three markers investigated between P. s. subflava and P. s. mutatrix. Otherwise leaning on our dating results, Prinia subflava subflava diverged from Prinia subflava mutatrix during the Pliocene epoch. -

Status and Seasonal Occurrence of Birds in Dhaka University Campus
Dhaka Univ. J. Biol. Sci. 25(1): 27‐37, 2016 (January) STATUS AND SEASONAL OCCURRENCE OF BIRDS IN DHAKA UNIVERSITY CAMPUS MST. FARHANA ARZUMAN BANU, MUNTASIR AKASH, GAWSIA WAHIDUNNESSA CHOWDHURY* AND MD. ANWAR HOSSAIN Department of Zoology, University of Dhaka, Dhaka 1000, Bangladesh Key words: Birds, Status, Seasonal occurrence Abstract A study was conducted on the status and seasonal occurrence of the birds in Dhaka University campus, from July, 2013 to February, 2014. During the study period, 54 species of birds belonging to 10 orders, 27 families and 47 genera were recorded. Of the recorded species of birds, 10 (19%) species were found as migrants and 44 (81%) as residents. Out of total species, 14 (26%) were identified as very common, 7 (13%) common, 10 (19%) uncommon, and 23 (43%) as rare. Of the resident species, 14 (25%) were very common residents, 26 (48%) common residents, 3 (6%) uncommon residents, 1 (2%) was rare resident and of the migrants, 7 (13%) were common winter visitors and 3 (6%) were found as uncommon winter visitors. From the study site, 28 (52%) species of passerine birds were observed and remaining 26 (48%) were non‐passerines. The highest number of birds was recorded in dry period/winter season (November ‐ February) and the lowest in wet period/monsoon (July ‐ October). Introduction Bird is a group of warm‐blooded, egg‐laying vertebrates of the class Aves having its body covered with feathers and forelimbs modified into wings(1,2). They have relatively large brain, keen sight and acute hearing, but little sense of smell. They share a common ancestry with reptiles and are believed to be evolved on the earth more than 150 million years ago(2‐4).