Evolution Unscathed: Darwin Devolves Argues on Weak Reasoning That Unguided Evolution Is a Destructive Force, Incapable of Innovation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Understanding the Intelligent Design Creationist Movement: Its True Nature and Goals
UNDERSTANDING THE INTELLIGENT DESIGN CREATIONIST MOVEMENT: ITS TRUE NATURE AND GOALS A POSITION PAPER FROM THE CENTER FOR INQUIRY OFFICE OF PUBLIC POLICY AUTHOR: BARBARA FORREST, Ph.D. Reviewing Committee: Paul Kurtz, Ph.D.; Austin Dacey, Ph.D.; Stuart D. Jordan, Ph.D.; Ronald A. Lindsay, J. D., Ph.D.; John Shook, Ph.D.; Toni Van Pelt DATED: MAY 2007 ( AMENDED JULY 2007) Copyright © 2007 Center for Inquiry, Inc. Permission is granted for this material to be shared for noncommercial, educational purposes, provided that this notice appears on the reproduced materials, the full authoritative version is retained, and copies are not altered. To disseminate otherwise or to republish requires written permission from the Center for Inquiry, Inc. Table of Contents Section I. Introduction: What is at stake in the dispute over intelligent design?.................. 1 Section II. What is the intelligent design creationist movement? ........................................ 2 Section III. The historical and legal background of intelligent design creationism ................ 6 Epperson v. Arkansas (1968) ............................................................................ 6 McLean v. Arkansas (1982) .............................................................................. 6 Edwards v. Aguillard (1987) ............................................................................. 7 Section IV. The ID movement’s aims and strategy .............................................................. 9 The “Wedge Strategy” ..................................................................................... -

Argumentation and Fallacies in Creationist Writings Against Evolutionary Theory Petteri Nieminen1,2* and Anne-Mari Mustonen1
Nieminen and Mustonen Evolution: Education and Outreach 2014, 7:11 http://www.evolution-outreach.com/content/7/1/11 RESEARCH ARTICLE Open Access Argumentation and fallacies in creationist writings against evolutionary theory Petteri Nieminen1,2* and Anne-Mari Mustonen1 Abstract Background: The creationist–evolutionist conflict is perhaps the most significant example of a debate about a well-supported scientific theory not readily accepted by the public. Methods: We analyzed creationist texts according to type (young earth creationism, old earth creationism or intelligent design) and context (with or without discussion of “scientific” data). Results: The analysis revealed numerous fallacies including the direct ad hominem—portraying evolutionists as racists, unreliable or gullible—and the indirect ad hominem, where evolutionists are accused of breaking the rules of debate that they themselves have dictated. Poisoning the well fallacy stated that evolutionists would not consider supernatural explanations in any situation due to their pre-existing refusal of theism. Appeals to consequences and guilt by association linked evolutionary theory to atrocities, and slippery slopes to abortion, euthanasia and genocide. False dilemmas, hasty generalizations and straw man fallacies were also common. The prevalence of these fallacies was equal in young earth creationism and intelligent design/old earth creationism. The direct and indirect ad hominem were also prevalent in pro-evolutionary texts. Conclusions: While the fallacious arguments are irrelevant when discussing evolutionary theory from the scientific point of view, they can be effective for the reception of creationist claims, especially if the audience has biases. Thus, the recognition of these fallacies and their dismissal as irrelevant should be accompanied by attempts to avoid counter-fallacies and by the recognition of the context, in which the fallacies are presented. -
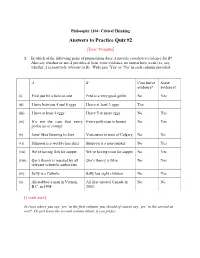
Answers to Practice Quiz #2
Philosophy 1104: Critical Thinking Answers to Practice Quiz #2 [Total: 50 marks] 1. In which of the following pairs of propositions does A provide conclusive evidence for B? Also say whether or not A provides at least some evidence, no matter how weak (i.e. say whether A is positively relevant to B). Write just „Yes‟ or „No‟ in each column provided. A B Conclusive Some evidence? evidence? (i) Fred just hit a hole-in-one Fred is a very good golfer No Yes (ii) I have between 4 and 6 eggs I have at least 3 eggs Yes (iii) I have at least 4 eggs I have 5 or more eggs No Yes (iv) It‟s not the case that every Every politician is honest No Yes politician is corrupt. (v) Janet likes listening to Jazz Vancouver is west of Calgary No No (vi) Simpson is a world-class skier Simpson is a non-smoker No Yes (vii) We‟re having fish for supper We‟re having trout for supper No Yes (viii) Qin‟s theory is rejected by all Qin‟s theory is false No Yes relevant scientific authorities (ix) Sally is a Catholic Sally has eight children No Yes (x) Ali stabbed a man in Vernon, Ali first entered Canada in No No B.C. in 1998. 2002. [1 mark each] In rows where you say ‘yes’ in the first column, you should of course say ‘yes’ in the second as well! Or just leave the second column blank, if you prefer. 2. Each of the following definitions is flawed in some way (each in just one way, I think, or at least one main one). -

Intelligent Design Is Not Science” Given by John G
An Outline of a lecture entitled, “Intelligent Design is not Science” given by John G. Wise in the Spring Semester of 2007: Slide 1 Why… … do humans have so much trouble with wisdom teeth? … is childbirth so dangerous and painful? Because a big, thinking brain is an advantage, and evolution is imperfect. Slide 2 Charles Darwin’s Revolutionary Idea His book changed the world. All life forms on this planet are related to each other through “Descent with modification over generations from a common ancestor”. Natural processes fully explain the biological connections between all life on the planet. Slide 3 Darwin’s Idea is Dangerous (from the book of the same title by Daniel Dennett) If we have evolution, we no longer need a Creator to create each and every species. Darwinism is dangerous because it infers that God did not directly and purposefully create us. It simply states that we evolved. Slide 4 Intelligent Design Attempts to Counter Darwin “Intelligent Design” has been proposed as way out of this dilemma. – Phillip Johnson, William Dembski, and Michael Behe (to name a few). They attempt to redefine science to encompass the supernatural as well as the natural world. They accept Darwin’s evolution, if an Intelligent Designer is (sometimes) substituted for natural selection. Design arguments are not new: − 1250 - St. Thomas Aquinas - first design argument − 1802 - Natural Theology - William Paley – perhaps the best(?) design argument Slide 5 What is Science? A specific way of understanding the natural world. Based on the idea that our senses give us accurate information about the Universe. -

Probability, Statistics, Evolution, and Intelligent Design
Probability, Statistics, Evolution, and Intelligent Design Peter Olofsson n the last decades, arguments against Darwinian evolution An ID Hypothesis Testing Challenge to Evolution have become increasingly sophisticated, replacing Cre- In his book The Design Inference, William Dembski introduces ationism by Intelligent Design (ID) and the book of Genesis I the “explanatory filter” as a device to rule out chance expla- by biochemistry and mathematics. As arguments claiming to nations and infer design of observed phenomena. The filter be based in probability and statistics are being used to justify also appears in his book No Free Lunch, where the description the anti-evolution stance, it may be of interest to readers of differs slightly. In essence, the filter is a variation on statistical CHANCE to investigate methods and claims of ID theorists. hypothesis testing with the main difference being that it aims at ruling out chance altogether, rather than just a specified Probability, Statistics, and Evolution null hypothesis. Once all chance explanations have been ruled The theory of evolution states in part that traits of organisms out, ‘design’ is inferred. Thus, in this context, design is merely are passed on to successive generations through genetic mate- viewed as the complement of chance. rial and that modifications in genetic material cause changes To illustrate the filter, Dembski uses the example of in appearance, ability, function, and survival of organisms. Nicholas Caputo, a New Jersey Democrat who was in charge Genetic changes that are advantageous to successful repro- of putting together the ballots in his county. Names were to be duction over time dominate and new species evolve. -

Does Intelligent Design Postulate a “Supernatural Creator?” Overview: No
Truth Sheet # 09-05 Does intelligent design postulate a “supernatural creator?” Overview: No. The ACLU, and many of its expert witnesses, have alleged that teaching the scientific theory of intelligent design (ID) is unconstitutional in all circumstances because it posits a “supernatural creator.” Yet actual statements from intelligent design theorists have made it clear that the scientific theory of intelligent design does not address metaphysical and religious questions such as the nature or identity of the designer. Firstly, the textbook being used in Dover, Of Pandas and People (Pandas ), makes it clear that design theory does not address religious or metaphysical questions, such as the nature or identity of the designer. Consider these two clear disclaimers from Pandas : "[T]he intelligent design explanation has unanswered questions of its own. But unanswered questions, which exist on both sides, are an essential part of healthy science; they define the areas of needed research. Questions often expose hidden errors that have impeded the progress of science. For example, the place of intelligent design in science has been troubling for more than a century. That is because on the whole, scientists from within Western culture failed to distinguish between intelligence, which can be recognized by uniform sensory experience, and the supernatural, which cannot. Today we recognize that appeals to intelligent design may be considered in science, as illustrated by current NASA search for extraterrestrial intelligence (SETI). Archaeology has pioneered the development of methods for distinguishing the effects of natural and intelligent causes. We should recognize, however, that if we go further, and conclude that the intelligence responsible for biological origins is outside the universe (supernatural) or within it, we do so without the help of science." 1 “[T]he concept of design implies absolutely nothing about beliefs normally associated with Christian fundamentalism, such as a young earth, a global flood, or even the existence of the Christian God. -

Irreducible Complexity (IC) Has Played a Pivotal Role in the Resurgence of the Creationist Movement Over the Past Two Decades
Irreducible incoherence and Intelligent Design: a look into the conceptual toolbox of a pseudoscience Abstract The concept of Irreducible Complexity (IC) has played a pivotal role in the resurgence of the creationist movement over the past two decades. Evolutionary biologists and philosophers have unambiguously rejected the purported demonstration of “intelligent design” in nature, but there have been several, apparently contradictory, lines of criticism. We argue that this is in fact due to Michael Behe’s own incoherent definition and use of IC. This paper offers an analysis of several equivocations inherent in the concept of Irreducible Complexity and discusses the way in which advocates of the Intelligent Design Creationism (IDC) have conveniently turned IC into a moving target. An analysis of these rhetorical strategies helps us to understand why IC has gained such prominence in the IDC movement, and why, despite its complete lack of scientific merits, it has even convinced some knowledgeable persons of the impending demise of evolutionary theory. Introduction UNTIL ITS DRAMATIC legal defeat in the Kitzmiller v. Dover case, Intelligent Design Creationism (IDC) had been one of the most successful pseudosciences of the past two decades, at least when measured in terms of cultural influence. It is interesting to explore the way this species of creationism had achieved this success, notwithstanding its periodic strategic setbacks as well as its complete lack of scientific merits. Of course, a full explanation would include religious, socio-political, and historical reasons; instead, in this paper, we will take a closer look at the conceptual toolbox and rhetorical strategies of the ID creationist. -

Clarity and Confusion
Book Reviews Clarity and confusion to overcome various antibiotics, such A review of as chloroquine, and humans have The Edge of Evolution: mutated to generate some measure of The Search for the resistance to malaria (e.g., sickle cell, Limits of Darwinism thalassemia). by Michael J. Behe Behe shows that all the cases of Free Press, New York, adaptation, in both Plasmodium and NY, 2007 humans, are due to breaking things, not creating new complex features. For example, chloroquine resistance Don Batten in Plasmodium is due to a fault in a transport protein that moves the poison his new book by Michael Behe, a into the organism’s vacuole. Behe Tfollow-up to Darwin’s Black Box likens the struggle to trench warfare, (DBB), has created somewhat of a where the defending forces will destroy storm amongst the faithful defenders their own bridge, or blow up a road, of Darwin such as Richard Dawkins, to impede the enemy’s advance. It is Jerry Coyne and Kenneth Miller. They not really an arms race, because in an arms race the opposing forces invent came out with all guns blazing to try overcoming other anti-malarials has to destroy the credibility of this book, new weapons, but the natural processes (‘evolution’) operating in Plasmodium only needed one mutation for each presumably hoping that their dismissive one. This seems like a reasonable vitriol would cause potential readers to and humans have not invented new weapons. explanation for the relative resilience skip reading it. Dawkins claimed that of chloroquine over time compared to Behe had completely departed from the Chloroquine resistance—a other anti-malarials. -

Intelligent Design: the Biochemical Challenge to Darwinian Evolution?
Christian Spirituality and Science Issues in the Contemporary World Volume 2 Issue 1 The Design Argument Article 2 2001 Intelligent Design: The Biochemical Challenge to Darwinian Evolution? Ewan Ward Avondale College Marty Hancock Avondale College Follow this and additional works at: https://research.avondale.edu.au/css Recommended Citation Ward, E., & Hancock, M. (2001). Intelligent design: The biochemical challenge to Darwinian evolution? Christian Spirituality and Science, 2(1), 7-23. Retrieved from https://research.avondale.edu.au/css/vol2/ iss1/2 This Article is brought to you for free and open access by the Avondale Centre for Interdisciplinary Studies in Science at ResearchOnline@Avondale. It has been accepted for inclusion in Christian Spirituality and Science by an authorized editor of ResearchOnline@Avondale. For more information, please contact [email protected]. Ward and Hancock: Intelligent Design Intelligent Design: The Biochemical Challenge to Darwinian Evolution? Ewan Ward and Marty Hancock Faculty of Science and Mathematics Avondale College “For since the creation of the world God’s invisible qualities – his eternal power and divine nature – have been clearly seen, being understood from what has been made, so that men are without excuse.” Romans 1:20 (NIV) ABSTRACT The idea that nature shows evidence of intelligent design has been argued by theologians and scientists for centuries. The most famous of the design argu- ments is Paley’s watchmaker illustration from his writings of the early 19th century. Interest in the concept of design in nature has recently had a resurgence and is often termed the Intelligent Design movement. Significant is the work of Michael Behe on biochemical systems. -

The College Student's Back to School Guide to Intelligent Design
Revised November, 2014 Part I: Letter of Introduction: Why this Student’s Guide? Part II: What is Intelligent Design? Part III: Answers to Your Professors’ 10 Most Common Misinformed Objections to Intelligent Design (1) Intelligent Design is Not Science (2) Intelligent Design is just a Negative Argument against Evolution (3) Intelligent Design Rejects All of Evolutionary Biology (4) Intelligent Design was Banned from Schools by the U.S. Supreme Court (5) Intelligent Design is Just Politics (6) Intelligent Design is a Science Stopper (7) Intelligent Design is “Creationism” and Based on Religion (8) Intelligent Design is Religiously Motivated (9) Intelligent Design Proponents Don’t Conduct or Publish Scientific Research (10) Intelligent Design is Refuted by the Overwhelming Evidence for Neo-Darwinian Evolution Part IV: Information About the Discovery Institute’s Summer Seminars on Intelligent Design COPYRIGHT © DISCOVERY INSTITUTE, 2014 — WWW.INTELLIGENTDESIGN.ORG PERMISSION GRANTED TO COPY AND DISTRIBUTE FOR NONPROFIT EDUCATIONAL PURPOSES. 2 Part I: Letter of Introduction: Why this Student’s Guide? Welcome to College, Goodbye to Intelligent Design? The famous Pink Floyd song that laments, “We don’t need no education / We don’t need no thought control,” is not just the rant of a rebellious mind; it is also a commentary on the failure of education to teach students how to think critically and evaluate both sides of controversial issues. Few scientists understood the importance of critical thinking better than Charles Darwin. When he first proposed his theory of evolution in Origin of Species in 1859, Darwin faced intense intellectual opposition from both the scientific community and the culture of his day. -

The Great Mutator Lution of Many of Which We Do Not Yet Fully Understand
living species, each of which had a slightly more advanced version of an eye. Behe’s Jerry A. Coyne novelty was to extend this argument to complex biochemical pathways, the evo- THE GREAT MUTATOR lution of many of which we do not yet fully understand. It was in the complexity of metabolism, blood clotting, and immu- THE EDGE OF EVOLUTION: this disclaimer is perfectly understand- nology that Behe claimed to have found THE SEARCH FOR THE LIMITS able. For in this department resides Mi- the hand of the Great Designer. OF DARWINISM chael Behe—that rara avis, a genuine The reviews of Darwin’s Black Box in By Michael J. Behe biologist who is also an advocate of “in- the scientific community were uniformly (Free Press, 320 pp., $28) telligent design.” And Lehigh University negative, for two reasons. First, we do does not wish to lose prospective stu- understand something about how these I. dents who bridle at the thought of study- pathways might have evolved in stepwise rowsing the websites of dif- ing miracles in their science courses. fashion, though we are as yet admittedly ferent colleges, a prospective Intelligent design, or ID, is a modern ignorant of many details. (It is harder to biology student finds an un- form of creationism cleverly constructed reconstruct the evolution of biochemi- usual statement on the page of to circumvent the many court decisions cal pathways than the evolution of or- the Department of Biological that have banned, on First Amendment ganisms themselves, because, unlike Sciences at Lehigh University. It begins: grounds, the teaching of religious views organisms, these pathways do not fos- B in the science classroom. -

Carroll on Behe-Intelligent Design
BOOKS ET AL. EVOLUTION proteins evolve and how proteins interact, and he completely ignores a huge amount of God as Genetic Engineer experimental data that directly contradicts his faulty premises. Unfortunately, these errors Sean B. Carroll are of a technical nature and will be difficult for lay readers, and even some scientists “The Lord hath delivered him into mine such as vertebrate classes). He also accepts (as (those unfamiliar with molecular biology and hands.” he has before) the 4.5-billion-year age of evolutionary genetics), to detect. Some people Earth and that we share a common ancestor will be hoodwinked. My goal here is to point hose are the words that Thomas Hux- with chimpanzees. That certainly won’t go out the critical flaws in Behe’s key arguments ley, Darwin’s confidant and staunchest over well in some camps. and to guide readers toward ally, purportedly murmured to a col- Behe also explores some ex- some references that illus- T The Edge of Evolution league as he rose to turn Bishop Samuel amples of Darwinian evolution trate why what he alleges to Wilberforce’s own words to his advantage and at the molecular level, including The Search for the be beyond the limits of rebut the bishop’s critique of Darwin’s theory an extensive treatment of the Limits of Darwinism Darwinian evolution falls at their legendary 1860 Oxford debate. They evolutionary “trench warfare” by Michael J. Behe well within its demonstrated are also the first words that popped into my fought between humans and Free Press, New York, 2007.