Genetic Comparison Between Victorian and Tasmanian Populations of Prasophyllum Correctum D.L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Flora Protetta (Misure Generali Di Conservazione Di Rete Natura 2000, Protezione Della Flora Spontanea) 2018 Dir
Flora protetta (Misure Generali di Conservazione di Rete Natura 2000, Protezione della Flora spontanea) 2018 Dir. Rete LR 2/77 Divisione Ordine Famiglia Taxon RER Sinonimie Habitat Natura 2000 Flora All. II-IV MGC spontanea Acarosporales Acarosporaceae Acarospora placodiiformis X Ascomycota Arthoniales Roccellaceae Ingaderia troglodytica Paralecanographa grumulosa X Lecanorales Cladoniaceae Cladonia spp. (group) X Entolomataceae Entoloma bloxamii X Agaricales Psathyrellaceae Psathyrella ammophila X Boletaceae Boletus dupainii X Boletales Paxillaceae Alpova rubescens X Basidiomycota Hymanochaetales Hymenochaetaceae Fomitiporia pseudopunctata Phellinus pseudopunctatus X Pezizales Pezizaceae Peziza pseudoammophila X Russulales Hericiaceae Hericium erinaceus X Xylariales Xylariaceae Poronia punctata X Bryales Bryaceae Bryum warneum Bryum oelandicum X Buxbaumiales Buxbaumiaceae Buxbaumia viridis X X Dicranales Leucobryaceae Leucobryum glaucum X Bryophyta Hypnales Amblystegiaceae Drepanocladus vernicosus Hamatocaulis vernicosus X X Othothrichales Othothrichaceae Orthotrichum rogeri X Pottiales Pottiaceae Tortula revolvens X Sphagnales Sphagnaceae Sphagnum spp. (group) X Diphasiastrum tristachyum Diphasium tristachyum X Diphasiastrum alpinum X Lycopodiales Lycopodiaceae Huperzia selago X Lycopodiophyta Lycopodium annotinum X Lycopodium clavatum X Selaginellales Selaginellaceae Selaginella selaginoides X Caldesia parnassifolia X X Alismataceae Baldellia ranunculoides X Alismatales Sagittaria sagittifolia X Hydrocharitaceae Stratiotes aloides -

Otanewainuku ED (Report Prepared on 13 August 2013)
1 NZFRI collection wish list for Otanewainuku ED (Report prepared on 13 August 2013) Fern Ally Isolepis cernua Lycopodiaceae Isolepis inundata Lycopodium fastigiatum Isolepis marginata Lycopodium scariosum Isolepis pottsii Psilotaceae Isolepis prolifera Tmesipteris lanceolata Lepidosperma australe Lepidosperma laterale Gymnosperm Schoenoplectus pungens Cupressaceae Schoenoplectus tabernaemontani Chamaecyparis lawsoniana Schoenus apogon Cupressus macrocarpa Schoenus tendo Pinaceae Uncinia filiformis Pinus contorta Uncinia gracilenta Pinus patula Uncinia rupestris Pinus pinaster Uncinia scabra Pinus ponderosa Hemerocallidaceae Pinus radiata Dianella nigra Pinus strobus Phormium cookianum subsp. hookeri Podocarpaceae Phormium tenax Podocarpus totara var. totara Iridaceae Prumnopitys taxifolia Crocosmia xcrocosmiiflora Libertia grandiflora Monocotyledon Libertia ixioides Agapanthaceae Watsonia bulbillifera Agapanthus praecox Juncaceae Alliaceae Juncus articulatus Allium triquetrum Juncus australis Araceae Juncus conglomeratus Alocasia brisbanensis Juncus distegus Arum italicum Juncus edgariae Lemna minor Juncus effusus var. effusus Zantedeschia aethiopica Juncus sarophorus Arecaceae Juncus tenuis var. tenuis Rhopalostylis sapida Luzula congesta Asparagaceae Luzula multiflora Asparagus aethiopicus Luzula picta var. limosa Asparagus asparagoides Orchidaceae Cordyline australis x banksii Acianthus sinclairii Cordyline banksii x pumilio Aporostylis bifolia Asteliaceae Corunastylis nuda Collospermum microspermum Diplodium alobulum Commelinaceae -

Liste Des Noms D'espèces D'orchidées De France Métropolitaine Utilisable Sur Le Site Orchisauvage
Date de mise à jour 16-avril-18 Liste des noms d'espèces d'orchidées de France métropolitaine utilisable sur le site Orchisauvage Il existe de nombreux synonymes de noms d'espèces utilisés par les botanistes et orchidophiles avec une évolution rapide. Afin de limiter les perturbations que cela crée pour les non spécialistes, le site Orchisauvage permet d'utiliser les synonymes les plus fréquents. En cas d'utilisation d'un synonyme, le nom retenu par la SFO, surligné en vert, sera celui affiché pour les observations accompagné d'un astérisque pour le repérer. Toutefois, le nom saisi sera toujours gardé dans la base de données. En plus des cartes de présence nationale pour chaque espèce, 17 cartes supplémentaires de regroupement d'espèces proches sont disponibles. Il s'agit des espèces indiquées "au sens large" surlignées en jaune. Nom d'orchidée pouvant être Nom valide ou retenu pour les Cartes supplémentaires de Nom vernaculaire utilisé en saisie restitutions regroupement d'espèces Aceras anthropophorum Orchis anthropophora Orchis homme pendu Anacamptis champagneuxii Anacamptis champagneuxii Orchis de Champagneux Orchis bouffon au sens large Anacamptis collina Anacamptis collina Orchis des collines Anacamptis coriophora Anacamptis coriophora Orchis punaise Orchis punaise au sens large Anacamptis indet. coriophora Anacamptis indéterminé de l’espèce coriophora Orchis punaise indéterminé Orchis punaise au sens large Anacamptis coriophora subsp. fragrans Anacamptis coriophora subsp. fragrans Orchis parfumé Orchis punaise au sens large Anacamptis coriophora subsp. martrinii Anacamptis coriophora subsp. martrinii Orchis de Martrin Orchis punaise au sens large Anacamptis laxiflora Anacamptis laxiflora Orchis à fleurs lâches Anacamptis longicornu Anacamptis longicornu Orchis à long éperon Anacamptis morio Anacamptis morio Orchis bouffon Orchis bouffon au sens large Anacamptis indet. -

Platanthera Chapmanii: Culture, Population Augmentation, and Mycorrhizal Associations
Platanthera chapmanii: culture, population augmentation, and mycorrhizal associations By Kirsten Poff, B.S. A Thesis In Plant and Soil Science Submitted to the Graduate Faculty of Texas Tech University in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE Approved Dr. Jyotsna Sharma Chair of Committee Dr. Scott Longing Dr. John Zak Dr. Mark Sheridan Dean of the Graduate School August, 2016 © 2016, Kirsten Poff Texas Tech University, Kirsten Poff, August 2016 ACKNOWLEDGEMENTS First I would like to thank my mentor and advisor, Dr. Jyotsna Sharma for all of her help and support. She has challenged and encouraged me throughout my program and the duration of this project. Thanks to her, I am light-years ahead of where I was two years ago. Texas Parks and Wildlife is also gratefully acknowledged for funding portions of this study. I also wish to express my gratitude to Dr. John Zak for his enthusiasm and for encouraging my love of microbes. I also gratefully thank Dr. Scott Longing for his advice, and constructive comments. I sincerely thank all three committee members for all the time and energy they have spent on me throughout the duration of my project. I gratefully acknowledge Dr. Jason Woodward for his encouragement and recommendations as well. I also acknowledge Dr. Cynthia McKenney and Mr. Russel Plowman for their support; I now have a passion for teaching, and a much better understanding of what it is like to teach college level courses. I want to also thank Mr. Robby Carlson for his time and technological assistance. -

Redalyc.ARE OUR ORCHIDS SAFE DOWN UNDER?
Lankesteriana International Journal on Orchidology ISSN: 1409-3871 [email protected] Universidad de Costa Rica Costa Rica BACKHOUSE, GARY N. ARE OUR ORCHIDS SAFE DOWN UNDER? A NATIONAL ASSESSMENT OF THREATENED ORCHIDS IN AUSTRALIA Lankesteriana International Journal on Orchidology, vol. 7, núm. 1-2, marzo, 2007, pp. 28- 43 Universidad de Costa Rica Cartago, Costa Rica Available in: http://www.redalyc.org/articulo.oa?id=44339813005 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative LANKESTERIANA 7(1-2): 28-43. 2007. ARE OUR ORCHIDS SAFE DOWN UNDER? A NATIONAL ASSESSMENT OF THREATENED ORCHIDS IN AUSTRALIA GARY N. BACKHOUSE Biodiversity and Ecosystem Services Division, Department of Sustainability and Environment 8 Nicholson Street, East Melbourne, Victoria 3002 Australia [email protected] KEY WORDS:threatened orchids Australia conservation status Introduction Many orchid species are included in this list. This paper examines the listing process for threatened Australia has about 1700 species of orchids, com- orchids in Australia, compares regional and national prising about 1300 named species in about 190 gen- lists of threatened orchids, and provides recommen- era, plus at least 400 undescribed species (Jones dations for improving the process of listing regionally 2006, pers. comm.). About 1400 species (82%) are and nationally threatened orchids. geophytes, almost all deciduous, seasonal species, while 300 species (18%) are evergreen epiphytes Methods and/or lithophytes. At least 95% of this orchid flora is endemic to Australia. -
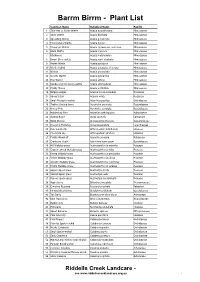
Botanical Name
Barrm Birrm - Plant List Common Name Botanical Name Family 1 Thin-leaf or Snake Wattle Acacia aculeatissima Mimosaceae 2 Silver Wattle Acacia dealbata Mimosaceae 3 Spreading Wattle Acacia genistifolia Mimosaceae 4 Ploughshare Wattle Acacia gunnii Mimosaceae 5 Cinnamon Wattle Acacia leprosa var. uninervia Mimosaceae 6 Black Wattle Acacia mearnsii Mimosaceae 7 Blackwood Acacia melanoxylon Mimosaceae 8 Dwarf Silver-wattle Acacia nano-dealbata Mimosaceae 9 Hedge Wattle Acacia paradoxa Mimosaceae 10 Wattle hybrid Acacia paradoxa x leprosa Mimosaceae 11 Wirilda Acacia provincialis Mimosaceae 12 Golden Wattle Acacia pycnantha Mimosaceae 13 Hop Wattle Acacia stricta Mimosaceae 14 Dandenong Cinnamon-wattle Acacia strictophylla Mimosaceae 15 Prickly Moses Acacia verticillata Mimosaceae 16 Bidgee-widgee Acaena novae-zelandiae Rosaceae 17 Sheep's Burr Acaena ovina Rosaceae 18 Small Mosquito-orchid Acianthus pusillus Orchidaceae 19 Trailing Ground-berry Acrotriche prostrata Epacridaceae 20 Honey Pots Acrotriche serrulata Epacridaceae 21 Maidenhair Fern Adiantum aethiopicum Adiantaceae 22 Austral Bugle Ajuga australis Lamiaceae 23 Black Sheoak Allocasuarina littoralis Casuarinaceae 24 Drooping Mistletoe Amyema pendula Loranthaceae 25 Pale Vanilla-lily Arthropodium milleflorum Liliaceae 26 Chocolate Lily Arthropodium strictum Liliaceae 27 Prickly Woodruff Asperula scoparia Rubiaceae 28 Cranberry Heath Astroloma humifusum Epacridaceae 29 Hill Wallaby-grass Austrodanthonia eriantha Poaceae 30 Copper-awned Wallaby-grass Austrodanthonia fulva Poaceae 31 -

JABG22P101 Barker
JOURNAL of the ADELAIDE BOTANIC GARDENS AN OPEN ACCESS JOURNAL FOR AUSTRALIAN SYSTEMATIC BOTANY flora.sa.gov.au/jabg Published by the STATE HERBARIUM OF SOUTH AUSTRALIA on behalf of the BOARD OF THE BOTANIC GARDENS AND STATE HERBARIUM © Board of the Botanic Gardens and State Herbarium, Adelaide, South Australia © Department of Environment, Water and Natural Resources, Government of South Australia All rights reserved State Herbarium of South Australia PO Box 2732 Kent Town SA 5071 Australia © 2008 Board of the Botanic Gardens & State Herbarium, Government of South Australia J. Adelaide Bot. Gard. 22 (2008) 101 –104 © 2008 Department for Environment & Heritage, Government of South Australia NOTES & SH ORT COMMUNICATIONS New combinations in Pterostylis and Caladenia and other name changes in the Orchidaceae of South Australia R.M. Barker & R.J. Bates State Herbarium of South Australia, Plant Biodiversity Centre, P.O. Box 2732, Kent Town, South Australia 5071 E-mail: [email protected] Abstract Combinations are provided in Pterostylis and Caladenia (Orchidaceae) for new species initially described in the segregate genera Arachnorchis, Bunochilus and Oligochaetochilus. Recircumscription of existing species has led to some new species being recognised for South Australia and Prasophyllum sp. West Coast (R.Tate AD96945167) is now known as Prasophyllum catenemum D.L.Jones. Introduction within Pterostylis2 R.Br. will not be adopted. Both In the past, when there have been disagreements genera in the wider sense are recognised as monophyletic between botanists about the level at which species (Hopper & Brown 2004; Jones & Clements 2002b) should be recognised, the arguments have not impinged and for the practical purpose of running Australia’s particularly on the outside community. -

Native Orchid Society South Australia
Journal of the Native Orchid Society of South Australia Inc PRINT POST APPROVED VOLUME 27 NO. 4 PP 54366200018 MAY 2003 NATIVE ORCHID SOCIETY OF SOUTH AUSTRALIA POST OFFICE BOX 565 UNLEY SOUTH AUSTRALIA 5061 The Native Orchid Society of South Australia promotes the conservation of orchids through the preservation of natural habitat and through cultivation. Except with the documented official representation from the Management Committee no person is authorised to represent the society on any matter. All native orchids are protected plants in the wild. Their collection without written Government permit is illegal. PRESIDENT: SECRETARY: Bodo Jensen Cathy Houston Telephone: 82430051 Work 8347 2005 Telephone: 8356 7356 VICE-PRESIDENT Bob Bates COMMITTEE Bill Dear Peter McCauley Malcolm Guy David Pettifor EDITOR: TREASURER David Hirst Iris Freeman 14 Beaverdale Avenue Windsor Gardens SA 5087 Telephone 8261 7998 E-mail [email protected] LIFE MEMBERS Mr R. Hargreaves Mr G. Carne Mr L. Nesbitt Mr R. Bates Mr R. Robjohns Mr R Shooter Mr D. Wells Registrar of Judges: Reg Shooter Trading Table: Judy Penney Field Trips & Conservation: Thelma Bridle Tel. 83844174 Tuber Bank Coordinator: Malcolm Guy Tel. 82767350 New Members Coordinator David Pettifor Tel. 0416 095 095 PATRON: Mr T.R.N. Lothian The Native Orchid Society of South Australia Inc. while taking all due care, take no responsibility for the loss, destruction or damage to any plants whether at shows, meetings or exhibits. Views or opinions expressed by authors of articles within this Journal do not necessarily reflect the views or opinions of the Management. We condones the reprint of any articles if acknowledgement is given. -

Ophrys Bertolonii, Ophrys Aurelia, Ophrys Romolinii
Orchid26 001-084 18.09.18 10:36 Page 53 www.orchidelforge.eu Natural. belges 94 (Orchid. 26) (2013): 53-60 Ophrys bertolonii, Ophrys aurelia, Ophrys romolinii par Pierre DELFORGE (*) Abstract. DELFORGE, P. - Ophrys bertolonii, Ophrys aurelia, Ophrys romolinii. The nomen- clatural controversy about Ophrys bertolonii and its solution are evoked. After the neotypi- fication of Ophrys bertolonii MORETTI 1823 by BAUMANN and KÜNKELE (1986) and the selection of an epitype by BAUMANN et al. (2002), Ophrys romolinii SOCA 1823 is definitively a posterior syn- onym of Ophrys bertolonii MORETTI 1823. Consequently, Ophrys aurelia P. DELFORGE, J. DEVILLERS- TERSCHUREN & P. DEVILLERS 1989 is not a posterior synonym of Ophrys bertolonii MORETTI 1823. The pres- ence of Ophrys bertolonii during two centuries in the surroundings of Genoa (Liguria, Italy), where it was described by BERTOLONI (1804) under the name of Ophrys speculum (nom. illegit., non LINK 1800), is confirmed. Key-Words: Orchidaceae; Ophrys bertolonii species group, Ophrys aurelia, Ophrys bertolonii, Ophrys romolinii. Nomenclature. Introduction Les controverses purement nomenclaturales ont été et sont malheureuse- ment assez fréquentes chez les Orchidées d’Europe, dont des taxons peu- vent, encore aujourd’hui, apparaître au même rang sous des noms qui diffè- rent selon les auteurs. Cette situation obscurcit et complique souvent leur abord comme leur étude et génère bien des difficultés notamment lorsqu’il faut prendre des dispositions légales pour leur conservation. En l’occurren- ce, quel nom donner, dans un arrêté, au taxon que l’on souhaite protéger ? La résolution définitive de ces problèmes agaçants est donc hautement sou- haitable et, lorsqu’elle intervient pour un taxon, elle ne devrait pas être négligée ni esquivée. -

BSBI News No. 54
B.S.B.I.NEWS April1990 Editedby R.Gwynn Ellis No.54 Dept.of Botany,National Museum ofWales Cardiff CF13NP f, Potamogeton friesii Potamogeton crispus L. Rupr. del. O.M. Srewarr @ rsso ,\drninist ration ADMINISTRATION HON. GENERAL SECRE-IARy (Generat Enquiries) Mrs Mary Briggs, M.B,E, 9 Arun Prospecr, PULBOROUGH, West Sussex RH20 IAL HON. TREASURER (Payment of Subscriptions and change of address) Mr Michael Walpole, 66 Outwoods Road, LOUGHBOROUGH, Leics LEll 3Ly (Please quote membership number on correspondence concerning membership or subscriptions - your membership number is on the address label of your mailings), HON. FIELD SECRETARY (Enquiries on Field fvleetings) Mr Roy Smith, 8 Salcey Close, SWANIiVICK, Derbys DE55 IHD SECRE*TARIES OF PERMANENT WORKING COMMIT'IEFS CONSERVATION (Acting Secretary) Mrs Elsa G. Wood, The Nurtons Field Centre, TINTERN, Chepstow, Gwent NP6 7NX MEETINGS: Mrs Ailsa Lee, 3, Rosliston Road, Stapenhill, BLIRTON-CN-TRENT, Staffordshire DEl5 9RJ PUBLICATIONS: Mr Arthur O. Chater, Dept. of Botany, fhe Natural Flisrory Museum, Cromwell Road, LONDON SWZ sBD RECORDS: Mr David J. N,lcCosh, l3 Cottesrnore Gardens, LONDON W8 5PR REGIONAL COMMITTEES 1989 . I99O SCOTLAND Dr P. i\4acpherson(lJon. Sec.), N,lr Fi.J. Noltie (Chairman), Nlrs Ni. Barron, Dr R.W.M. Corner, Dr Nl.G.B. Hughes, Dr H. I-ang, Nliss J. lvluscott, i\,lr M.l,{. Scott, Dr R.A.H. Smith, Mr N.F. Stewart, Mr,A. l\'lcG. Stirling, Nir B.lJ. Thornpson, l\{r J. Winham. Represenrative on Council: Mr fl.J. Nolrie. IRELAND Dr T.F.G. Curtis (Chairman), Mr S. Reesley, tulr D.A. -

Biodiversity Summary for NRM Regions Species List
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

Muelleria 18: 99Ð109 (2003)
Muelleria 18: 99–109 (2003) A revisionary treatment of four species of Prasophyllum R.Br. (Orchidaceae) loosely related to P. correctum D.L.Jones David L. Jones Centre for Plant Biodiversity Research, Australian National Herbarium, G.P.O. Box 1600, Canberra, Australian Capital Territory, 2601, Australia. [email protected] Abstract The taxonomy of a small group of species loosely related to Prasophyllum correctum (Orchidaceae) is resolved. Four species are recognised in the group - P. correctum D.L.Jones, P. bagoensis D.L.Jones and two species newly described here (P. c rebriflorum D.L.Jones and P. incorrectum D.L.Jones). The taxonomic significance of the latter new species was resolved by an independent group of researchers (Orthia et al.) using a molecular technique, the results of this study being reported in an accompanying paper in this issue. Subsequent re-examination of morphological characters supports these findings. Keywords: Prasophyllum,new species, P. correctum, P. bagoensis, P. c re b riflorum, P. incorrectum, Victoria, Tasmania, Australia. Introduction Prasophyllum R.Br. is a complex genus of Australian and New Zealand Orchidaceae which presents difficulties in identification for taxonomists, ecologists and orchid enthusiasts, mainly because of general similarity between many taxa and the difficulty of defining unique characters which can be used as a ready means of identification. Prasophyllum is the subject of continuing studies which have resulted in the description of new species (Bates 1989b, 1990; Jones 1991, 1994a, 1994b, 1996b, 1997), a review of Tasmanian species (Jones 1998) and the resolution of various complexes (Bates 1989a; Jones 1996a; Jones & Clements 1996).