Shakya Et Al., 2017
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Assessing Conservation Status of Resident and Migrant Birds on Hispaniola with Mist-Netting
Assessing conservation status of resident and migrant birds on Hispaniola with mist-netting John D. Lloyd, Christopher C. Rimmer and Kent P. McFarland Vermont Center for Ecostudies, Norwich, VT, United States ABSTRACT We analyzed temporal trends in mist-net capture rates of resident (n D 8) and overwintering Nearctic-Neotropical migrant (n D 3) bird species at two sites in montane broadleaf forest of the Sierra de Bahoruco, Dominican Republic, with the goal of providing quantitative information on population trends that could inform conservation assessments. We conducted sampling at least once annually during the winter months of January–March from 1997 to 2010. We found evidence of declines in capture rates for three resident species, including one species endemic to Hispaniola. Capture rate of Rufous-throated Solitaire (Myadestes genibarbis) declined by 3.9% per year (95% CL D 0%, 7.3%), Green-tailed Ground-Tanager (Microligea palustris) by 6.8% (95% CL D 3.9%, 8.8%), and Greater Antillean Bullfinch (Loxigilla violacea) by 4.9% (95% CL D 0.9%, 9.2%). Two rare and threatened endemics, Hispaniolan Highland-Tanager (Xenoligea montana) and Western Chat-Tanager (Calyptophilus tertius), showed statistically significant declines, but we have low confidence in these findings because trends were driven by exceptionally high capture rates in 1997 and varied between sites. Analyses that excluded data from 1997 revealed no trend in capture rate over the course of the study. We found no evidence of temporal trends in capture rates for any other residents or Nearctic-Neotropical migrants. We do not know the causes of the observed declines, nor can we conclude that these declines are not a purely Submitted 12 September 2015 local phenomenon. -

Species Limits in Some Philippine Birds Including the Greater Flameback Chrysocolaptes Lucidus
FORKTAIL 27 (2011): 29–38 Species limits in some Philippine birds including the Greater Flameback Chrysocolaptes lucidus N. J. COLLAR Philippine bird taxonomy is relatively conservative and in need of re-examination. A number of well-marked subspecies were selected and subjected to a simple system of scoring (Tobias et al. 2010 Ibis 152: 724–746) that grades morphological and vocal differences between allopatric taxa (exceptional character 4, major 3, medium 2, minor 1; minimum score 7 for species status). This results in the recognition or confirmation of species status for (inverted commas where a new English name is proposed) ‘Philippine Collared Dove’ Streptopelia (bitorquatus) dusumieri, ‘Philippine Green Pigeon’ Treron (pompadora) axillaris and ‘Buru Green Pigeon’ T. (p.) aromatica, Luzon Racquet-tail Prioniturus montanus, Mindanao Racquet-tail P. waterstradti, Blue-winged Raquet-tail P. verticalis, Blue-headed Raquet-tail P. platenae, Yellow-breasted Racquet-tail P. flavicans, White-throated Kingfisher Halcyon (smyrnensis) gularis (with White-breasted Kingfisher applying to H. smyrnensis), ‘Northern Silvery Kingfisher’ Alcedo (argentata) flumenicola, ‘Rufous-crowned Bee-eater’ Merops (viridis) americanus, ‘Spot-throated Flameback’ Dinopium (javense) everetti, ‘Luzon Flameback’ Chrysocolaptes (lucidus) haematribon, ‘Buff-spotted Flameback’ C. (l.) lucidus, ‘Yellow-faced Flameback’ C. (l.) xanthocephalus, ‘Red-headed Flameback’ C. (l.) erythrocephalus, ‘Javan Flameback’ C. (l.) strictus, Greater Flameback C. (l.) guttacristatus, ‘Sri Lankan Flameback’ (Crimson-backed Flameback) Chrysocolaptes (l.) stricklandi, ‘Southern Sooty Woodpecker’ Mulleripicus (funebris) fuliginosus, Visayan Wattled Broadbill Eurylaimus (steerii) samarensis, White-lored Oriole Oriolus (steerii) albiloris, Tablas Drongo Dicrurus (hottentottus) menagei, Grand or Long-billed Rhabdornis Rhabdornis (inornatus) grandis, ‘Visayan Rhabdornis’ Rhabdornis (i.) rabori, and ‘Visayan Shama’ Copsychus (luzoniensis) superciliaris. -

A Revision of the Mexican Piculus (Picidae) Complex
THE WILSON BULLETIN A QUARTERLY MAGAZINE OF ORNITHOLOGY Published by the Wilson Ornithological Society VOL. 90, No. 2 JUNE 1978 PAGES 159-334 WilsonBdl., 90(Z), 1978, pp. 159-181 A REVISION OF THE MEXICAN PZCULUS (PICIDAE) COMPLEX LUIS F. BAPTISTA The neotropical woodpeckers of the genus Piculus are closely related to the flickers (Colaptes) (Short 1972). P icu 1us species range from Mexico to southern Brazil, Paraguay, Peru (Ridgway 1914) and Argentina (Salvin and Godman 1892). Peters (1948) lists 46 taxa (9 species and their sub- species) of which 20 are subspecies of Piculus rubiginosus, the most widely distributed species. The latter ranges from southern Veracruz to the north- western provinces of Jujuy, Salta, and Tucuman in Argentina (Peters 1948). Compared with other picids, this genus is generally poorly represented in museum collections. It is possible that they are not as rare as they seem, but being rather silent and secretive birds and difficult to distinguish from the associated vegetation due to their cryptic green coloration, are easily passed unnoticed by collectors in the field. A difference of opinion exists among taxonomists regarding the status of several of the Mexican forms. Two species complexes are recognized in the Mexican check-list (Miller et al. 1957). The Piculus auricularti complex is reported by these authors as consisting of 2 subspecies: sonoriensis known only from the type series of 3 birds taken at Ranch0 Santa Barbara, Sonora, and the nominate race auricularis recorded as ranging from Sinaloa south to Guerrero. They point out the uncertain status of the form sonorien- sis, stating that additional material is needed to substantiate it. -

Patterns of Co-Occurrence in Woodpeckers and Nocturnal Cavity-Nesting Owls Within an Idaho Forest
VOLUME 13, ISSUE 1, ARTICLE 18 Scholer, M. N., M. Leu, and J. R. Belthoff. 2018. Patterns of co-occurrence in woodpeckers and nocturnal cavity-nesting owls within an Idaho forest. Avian Conservation and Ecology 13(1):18. https://doi.org/10.5751/ACE-01209-130118 Copyright © 2018 by the author(s). Published here under license by the Resilience Alliance. Research Paper Patterns of co-occurrence in woodpeckers and nocturnal cavity- nesting owls within an Idaho forest Micah N. Scholer 1, Matthias Leu 2 and James R. Belthoff 1 1Department of Biological Sciences and Raptor Research Center, Boise State University, Boise, Idaho, USA, 2Biology Department, College of William and Mary, Williamsburg, Virginia, USA ABSTRACT. Few studies have examined the patterns of co-occurrence between diurnal birds such as woodpeckers and nocturnal birds such as owls, which they may facilitate. Flammulated Owls (Psiloscops flammeolus) and Northern Saw-whet Owls (Aegolius acadicus) are nocturnal, secondary cavity-nesting birds that inhabit forests. For nesting and roosting, both species require natural cavities or, more commonly, those that woodpeckers create. Using day and nighttime broadcast surveys (n = 150 locations) in the Rocky Mountain biogeographic region of Idaho, USA, we surveyed for owls and woodpeckers to assess patterns of co-occurrence and evaluated the hypothesis that forest owls and woodpeckers co-occurred more frequently than expected by chance because of the facilitative nature of their biological interaction. We also examined co-occurrence patterns between owl species to understand their possible competitive interactions. Finally, to assess whether co-occurrence patterns arose because of species interactions or selection of similar habitat types, we used canonical correspondence analysis (CCA) to examine habitat associations within this cavity-nesting bird community. -

Dominican Republic Endemics of Hispaniola II 1St February to 9Th February 2021 (9 Days)
Dominican Republic Endemics of Hispaniola II 1st February to 9th February 2021 (9 days) Palmchat by Adam Riley Although the Dominican Republic is perhaps best known for its luxurious beaches, outstanding food and vibrant culture, this island has much to offer both the avid birder and general naturalist alike. Because of the amazing biodiversity sustained on the island, Hispaniola ranks highest in the world as a priority for bird protection! This 8-day birding tour provides the perfect opportunity to encounter nearly all of the island’s 32 endemic bird species, plus other Greater Antillean specialities. We accomplish this by thoroughly exploring the island’s variety of habitats, from the evergreen and Pine forests of the Sierra de Bahoruco to the dry forests of the coast. Furthermore, our accommodation ranges from remote cabins deep in the forest to well-appointed hotels on the beach, each with its own unique local flair. Join us for this delightful tour to the most diverse island in the Caribbean! RBL Dominican Republic Itinerary 2 THE TOUR AT A GLANCE… THE ITINERARY Day 1 Arrival in Santo Domingo Day 2 Santo Domingo Botanical Gardens to Sabana del Mar (Paraiso Caño Hondo) Day 3 Paraiso Caño Hondo to Santo Domingo Day 4 Salinas de Bani to Pedernales Day 5 Cabo Rojo & Southern Sierra de Bahoruco Day 6 Cachote to Villa Barrancoli Day 7 Northern Sierra de Bahoruco Day 8 La Placa, Laguna Rincon to Santo Domingo Day 9 International Departures TOUR ROUTE MAP… RBL Dominican Republic Itinerary 3 THE TOUR IN DETAIL… Day 1: Arrival in Santo Domingo. -

Bird List Column A: We Should Encounter (At Least a 90% Chance) Column B: May Encounter (About a 50%-90% Chance) Column C: Possible, but Unlikely (20% – 50% Chance)
THE PHILIPPINES Prospective Bird List Column A: we should encounter (at least a 90% chance) Column B: may encounter (about a 50%-90% chance) Column C: possible, but unlikely (20% – 50% chance) A B C Philippine Megapode (Tabon Scrubfowl) X Megapodius cumingii King Quail X Coturnix chinensis Red Junglefowl X Gallus gallus Palawan Peacock-Pheasant X Polyplectron emphanum Wandering Whistling Duck X Dendrocygna arcuata Eastern Spot-billed Duck X Anas zonorhyncha Philippine Duck X Anas luzonica Garganey X Anas querquedula Little Egret X Egretta garzetta Chinese Egret X Egretta eulophotes Eastern Reef Egret X Egretta sacra Grey Heron X Ardea cinerea Great-billed Heron X Ardea sumatrana Purple Heron X Ardea purpurea Great Egret X Ardea alba Intermediate Egret X Ardea intermedia Cattle Egret X Ardea ibis Javan Pond-Heron X Ardeola speciosa Striated Heron X Butorides striatus Yellow Bittern X Ixobrychus sinensis Von Schrenck's Bittern X Ixobrychus eurhythmus Cinnamon Bittern X Ixobrychus cinnamomeus Black Bittern X Ixobrychus flavicollis Black-crowned Night-Heron X Nycticorax nycticorax Western Osprey X Pandion haliaetus Oriental Honey-Buzzard X Pernis ptilorhynchus Barred Honey-Buzzard X Pernis celebensis Black-winged Kite X Elanus caeruleus Brahminy Kite X Haliastur indus White-bellied Sea-Eagle X Haliaeetus leucogaster Grey-headed Fish-Eagle X Ichthyophaga ichthyaetus ________________________________________________________________________________________________________ WINGS ● 1643 N. Alvernon Way Ste. 109 ● Tucson ● AZ ● 85712 ● www.wingsbirds.com -

The Generic Distinction of Pied Woodpeckers
THE GENERIC DISTINCTION OF PIED WOODPECKERS M. RALPH BROWNING, 170 JacksonCreek Drive, Jacksonville,Oregon 97530 ABSTRACT: The ten speciesof New World four-toedwoodpeckers (scalaris, nuttallii, pubescens, villosus, stricklandi, arizonae, borealis, albolarvatus, lignarius,and m ixtusand the two borealthree-toed species (arcticus and tridactylus), currentlycombined in the genusPicoides, differ, in additionto the numberof toes,in modificationsof the skull,ribs, the belly of the pubo-ischio-femoralismuscle, head plumage,and behavior. I recommendthat the genericname Dryobates be reinstituted for the New World four-toedwoodpeckers. There are three generalmorphological groups of pied woodpeckers,a groupof nine four-toedspecies of the New World, a groupof 22 four-toed speciesof the Old World, and a groupof two three-toedspecies straddling bothregions. ! referto thesegroups of piedwoodpeckers beyond as the New World,Old World,and three-toedgroups. The three-toedspecies have long beenin the genusPicoides Lac•p•de, 1799, but the four-toedgroups have been combinedat the genericlevel in differentways. All four-toedpied woodpeckerswere long includedin the genusDryobates Boie, 1826, later changed to Dendrocopos Koch, 1816 an earlier name (Voous 1947, A.O.U. 1947, Peters 1948). Despite the differencein number of toes, Dendrocoposwas combined with Picoidesbecause of generalsimilarities in anatomy (Delacour 1951, Short 1971a), plumage and behavior (Short 1974a), and vocalizations(Winkler and Short 1978). The A.O.U (1976) followedthis mergerof the genera.On the basisof skeletalcharacters Rea (1983) was skepticalof the merger,but he did not providedetails. On the otherhand, Ouellet(1977), concludingthat the two generadiffer in external morphologyand some behaviors and vocalizations, separated the Old World four-toedwoodpeckers in Dendrocoposand three-toedand New World four-toedwoodpeckers in Picoides.The A.O.U. -

(Dendrocopos Syriacus) and Middle Spotted Woodpecker (Dendrocoptes Medius) Around Yasouj City in Southwestern Iran
Archive of SID Iranian Journal of Animal Biosystematics (IJAB) Vol.15, No.1, 99-105, 2019 ISSN: 1735-434X (print); 2423-4222 (online) DOI: 10.22067/ijab.v15i1.81230 Comparison of nest holes between Syrian Woodpecker (Dendrocopos syriacus) and Middle Spotted Woodpecker (Dendrocoptes medius) around Yasouj city in Southwestern Iran Mohamadian, F., Shafaeipour, A.* and Fathinia, B. Department of Biology, Faculty of Science, Yasouj University, Yasouj, Iran (Received: 10 May 2019; Accepted: 9 October 2019) In this study, the nest-cavity characteristics of Middle Spotted and Syrian Woodpeckers as well as tree characteristics (i.e. tree diameter at breast height and hole measurements) chosen by each species were analyzed. Our results show that vertical entrance diameter, chamber vertical depth, chamber horizontal depth, area of entrance and cavity volume were significantly different between Syrian Woodpecker and Middle Spotted Woodpecker (P < 0.05). The average tree diameter at breast height and nest height between the two species was not significantly different (P > 0.05). For both species, the tree diameter at breast height and nest height did not significantly correlate. The directions of nests’ entrances were different in the two species, not showing a preferentially selected direction. These two species chose different habitats with different tree coverings, which can reduce the competition between the two species over selecting a tree for hole excavation. Key words: competition, dimensions, nest-cavity, primary hole nesters. INTRODUCTION Woodpeckers (Picidae) are considered as important excavator species by providing cavities and holes to many other hole-nesting species (Cockle et al., 2011). An important part of habitat selection in bird species is where to choose a suitable nest-site (Hilden, 1965; Stauffer & Best, 1982; Cody, 1985). -

The Gambia: a Taste of Africa, November 2017
Tropical Birding - Trip Report The Gambia: A Taste of Africa, November 2017 A Tropical Birding “Chilled” SET DEPARTURE tour The Gambia A Taste of Africa Just Six Hours Away From The UK November 2017 TOUR LEADERS: Alan Davies and Iain Campbell Report by Alan Davies Photos by Iain Campbell Egyptian Plover. The main target for most people on the tour www.tropicalbirding.com +1-409-515-9110 [email protected] p.1 Tropical Birding - Trip Report The Gambia: A Taste of Africa, November 2017 Red-throated Bee-eaters We arrived in the capital of The Gambia, Banjul, early evening just as the light was fading. Our flight in from the UK was delayed so no time for any real birding on this first day of our “Chilled Birding Tour”. Our local guide Tijan and our ground crew met us at the airport. We piled into Tijan’s well used minibus as Little Swifts and Yellow-billed Kites flew above us. A short drive took us to our lovely small boutique hotel complete with pool and lovely private gardens, we were going to enjoy staying here. Having settled in we all met up for a pre-dinner drink in the warmth of an African evening. The food was delicious, and we chatted excitedly about the birds that lay ahead on this nine- day trip to The Gambia, the first time in West Africa for all our guests. At first light we were exploring the gardens of the hotel and enjoying the warmth after leaving the chilly UK behind. Both Red-eyed and Laughing Doves were easy to see and a flash of colour announced the arrival of our first Beautiful Sunbird, this tiny gem certainly lived up to its name! A bird flew in landing in a fig tree and again our jaws dropped, a Yellow-crowned Gonolek what a beauty! Shocking red below, black above with a daffodil yellow crown, we were loving Gambian birds already. -

Federal Register/Vol. 81, No. 200/Monday, October 17, 2016
Federal Register / Vol. 81, No. 200 / Monday, October 17, 2016 / Proposed Rules 71457 for the relevant maintenance period in attainment of the 2008 ozone NAAQS Technology Transfer and Advancement with mobile source emissions at the through 2030. Finally, EPA finds Act of 1995 (15 U.S.C. 272 note) because levels of the MVEBs. adequate and is proposing to approve application of those requirements would the newly-established 2020 and 2030 be inconsistent with the CAA; and C. What is a safety margin? MVEBs for the Cleveland area. • Does not provide EPA with the A ‘‘safety margin’’ is the difference discretionary authority to address, as VII. Statutory and Executive Order between the attainment level of appropriate, disproportionate human Reviews emissions (from all sources) and the health or environmental effects, using projected level of emissions (from all Under the CAA, redesignation of an practicable and legally permissible sources) in the maintenance plan. As area to attainment and the methods, under Executive Order 12898 noted in Table 11, the emissions in the accompanying approval of a (59 FR 7629, February 16, 1994). Cleveland area are projected to have maintenance plan under section In addition, the SIP is not approved safety margins of 117.22 TPSD for NOX 107(d)(3)(E) are actions that affect the to apply on any Indian reservation land and 28.48 TPSD for VOC in 2030 (the status of a geographical area and do not or in any other area where EPA or an total net change between the attainment impose any additional regulatory Indian tribe has demonstrated that a year, 2014, emissions and the projected requirements on sources beyond those tribe has jurisdiction. -
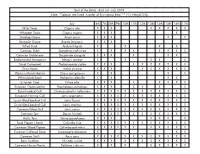
Best of the Baltic - Bird List - July 2019 Note: *Species Are Listed in Order of First Seeing Them ** H = Heard Only
Best of the Baltic - Bird List - July 2019 Note: *Species are listed in order of first seeing them ** H = Heard Only July 6th 7th 8th 9th 10th 11th 12th 13th 14th 15th 16th 17th Mute Swan Cygnus olor X X X X X X X X Whopper Swan Cygnus cygnus X X X X Greylag Goose Anser anser X X X X X Barnacle Goose Branta leucopsis X X X Tufted Duck Aythya fuligula X X X X Common Eider Somateria mollissima X X X X X X X X Common Goldeneye Bucephala clangula X X X X X X Red-breasted Merganser Mergus serrator X X X X X Great Cormorant Phalacrocorax carbo X X X X X X X X X X Grey Heron Ardea cinerea X X X X X X X X X Western Marsh Harrier Circus aeruginosus X X X X White-tailed Eagle Haliaeetus albicilla X X X X Eurasian Coot Fulica atra X X X X X X X X Eurasian Oystercatcher Haematopus ostralegus X X X X X X X Black-headed Gull Chroicocephalus ridibundus X X X X X X X X X X X X European Herring Gull Larus argentatus X X X X X X X X X X X X Lesser Black-backed Gull Larus fuscus X X X X X X X X X X X X Great Black-backed Gull Larus marinus X X X X X X X X X X X X Common/Mew Gull Larus canus X X X X X X X X X X X X Common Tern Sterna hirundo X X X X X X X X X X X X Arctic Tern Sterna paradisaea X X X X X X X Feral Pigeon ( Rock) Columba livia X X X X X X X X X X X X Common Wood Pigeon Columba palumbus X X X X X X X X X X X Eurasian Collared Dove Streptopelia decaocto X X X Common Swift Apus apus X X X X X X X X X X X X Barn Swallow Hirundo rustica X X X X X X X X X X X Common House Martin Delichon urbicum X X X X X X X X White Wagtail Motacilla alba X X -

K Here for the Full Trip Report
Capped Langur , Small Pratincole , WhiteWhite----wingedwinged Ducks , Gaur , Sultan Tit , Great Hornbill andand Pied Falconet ; Nameri Here a couple of ducks had now turned up , and while we were watching them , a Gaur suddenly came out of the forest for a drink and some fresh grass from the meadow surrounding the lake. We also saw several Wild Boars and a small group of Northern Red Muntjacs here , not to mention a Great Hornbill , Sultan Tits and a small party of Scarlet Minivets. No doubt this was a fantastic place , and there is no telling what could have been seen if more time had been spent here. Back in the camp we enjoyed yet another good meal , before driving a bit up river where this afternoons boat ride was to begin. We didn’t really see all that many birds while rafting on the river , but even so it was a nice experience to watch the beautiful landscape pass by in a leisurely pace – not exactly white water rafting this! Of course , there were a few avian highlights as well , including lots of Small Pratincoles , a couple of Crested Kingfisher and some nice River Lapwings , but we somehow managed to dip out on Ibisbill. We walked back to the camp as the sun was setting , but didn’t add anything new to our list , though a couple of Brown Hawk Owls put on quite a show for Erling , who was the first one to get back. We tried again with some spotlighting in the evening , and heard a calling Oriental Scops Owl not to far from the road but still impossible to see.