Genetic Verification of Multiple Paternity in Two Free-Ranging Isolated Populations of African Wild Dogs (Lycaon Pictus)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
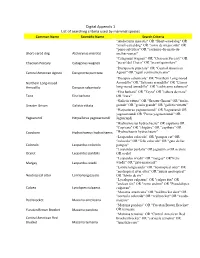
Digital Appendix 1 List of Searching Criteria Used by Mammal Species
Digital Appendix 1 List of searching criteria used by mammal species Common Name Scientific Name Search Criteria "Atelocynus microtis" OR "Short-eared dog" OR "small-eared dog" OR "zorro de oreja corta" OR "perro selvático" OR "cachorro-do-mato-de- Short-eared dog Atelocynus microtis orelhas-curtas" "Catagonus wagneri" OR "Chacoan Peccary" OR Chacoan Peccary Catagonus wagneri "pecarí del Chaco" OR "pecarí quimilero" “Dasyprocta punctate” OR "Central American Central American Agouti Dasyprocta punctata Agouti" OR "agutí centroamericano" “Dasypus sabanicola” OR "Northern Long-nosed Northern Long-nosed Armadillo" OR "Savanna armadillo" OR "Llanos Armadillo Dasypus sabanicola long-nosed armadillo" OR "cachicamo sabanero" “Eira barbara” OR "Tayra" OR "cabeza de mate" Taira Eira barbara OR "irara" “Galictis vittata” OR "Greater Grison" OR "furão- Greater Grison Galictis vittata grande" OR "grisón grande" OR "galictis vittatta" “Herpailurus yagouaroundi” OR Yaguarundi OR yagouaroundi OR "Puma yagouaroundi" OR Yaguarundi Herpailurus yagouaroundi Jaguarundi "Hydrochoerus hydrochaeris" OR capybara OR "Capivara" OR "chigüire" OR "capibara" OR Capybara Hydrochoerus hydrochaeris "Hydrochaeris hydrochaeris" “Leopardus colocolo” OR "pampas cat" OR "colocolo" OR "felis colocolo" OR "gato de las Colocolo Leopardus colocolo pampas" "Leopardus pardalis" OR jaguatirica OR ocelote Ocelot Leopardus pardalis OR ocelot “Leopardus wiedii” OR "margay" OR"Felis Margay Leopardus wiedii wiedii" OR "gato-maracajá" “Lontra longicaudis” OR "neotropical otter" OR "neotropical -

Galictis Cuja Molina, 1782) As Host of Dioctophyme Renale Goeze, 1782 Furão Pequeno (Galictis Cuja Molina, 1782) Como Hospedeiro De Dioctophyme Renale Goeze, 1782
ANIMAL PARASITOLOGY / SCIENTIFIC COMMUNICATION DOI: 10.1590/1808-1657000312016 Lesser Grison (Galictis cuja Molina, 1782) as host of Dioctophyme renale Goeze, 1782 Furão Pequeno (Galictis cuja Molina, 1782) como hospedeiro de Dioctophyme renale Goeze, 1782 Daniela Pedrassani1*, Mayana Worm1, Jéssica Drechmer1, Margareth Cristina Iazzetti Santos1 ABSTRACT: The Dioctophyme renale is a helminth parasite RESUMO: O Dioctophyme renale é um helminto parasita renal of the kidney usually seen in domestic and wild carnivores and observado normalmente em carnívoros domésticos e silvestres e rarely in human beings. This is a report about the parasitism excepcionalmente em seres humanos. Relata-se o parasitismo por D. of D. renale found in the kidney of two roadkill lesser grisons renale em rim de dois furões pequenos (Galictis cuja) encontrados (Galictis cuja) in the North of the state of Santa Catarina, mortos por atropelamento no Norte do estado de Santa Catarina, Brazil. The report of this parasitism in this species is important Brasil. Relatar esse parasitismo nessa espécie é importante, para to complement the records about this native carnivore as a que se possam somar dados relativos a participação deste carnívoro contributor in the epidemiologic chain while host/disseminator nativo na cadeia epidemiológica como hospedeiro/ veiculador desse of this helminth with zoonotic potential. helminto com potencial zoonótico. KEYWORDS: Dioctophyma; wild animal; mustelids; roadkill; PALAVRAS-CHAVE: Dioctophyma; animal silvestre; mustelí- kidney parasitism. deo; atropelamento em rodovia; parasitismo renal. 1Universidade do Contestado (UnC) – Canoinhas (SC), Brazil. *Corresponding author: [email protected] Received on: 04/22/2016. Accepted on: 09/12/2017 Arq. Inst. Biol., v.84, 1-4, e0312016, 2017 1 D. -

KEIDEL Itinerary
GILDEA, KEIDEL AND SCHULTZ ITINERARY SOUTH AFRICA - DECEMBER 2014 DAY BY DAY DAY 1 (SUNDAY 7 DECEMBER) Pick up your rental car and head towards Lesedi Cultural Village. Your accommodation on the 6th had not been confirmed yet, but more than likely the fire and ice hotel in Melrose Arch, in which case the following directions are suitable. From Johannesburg take the M1 north towards Pretoria, (the highway right next to Melrose Arch) and then turn west onto the N1 at the Woodmead interchange, following signs to Bloemfontein (Roughly 10km after you get onto the highway). At the Malabongwe drive off-ramp (roughly another 10 km), turn off and then turn right onto Malabongwe drive (R512), proceed for 40 kms along the R512, Lesedi is clearly marked on the left-hand side of the rd. The Lesedi Staff will meet you on arrival. Start your journey at the Ndebele village with an introduction to the cultural experience preceded by a multi-media presentation on the history and origins of South Africa’s rainbow nation. Then, enjoy a guided tour of the other four ethnic homesteads – Zulu, Basotho, Xhosa and Pedi. As the sun sets over the African bush, visit the Boma for a very interactive affair of traditional singing and dancing, which depict stories dating back to the days of their ancestors. Dine in the Nyama Choma Restaurant, featuring ethnic dishes, a fusion of Pan African cuisine all complemented by warm, traditional service. Lesedi African Lodge and Cultural Village Kalkheuwel, Broederstroom R512, Lanseria, 1748 [t] 087 940 9933 [m] 071 507 1447 DAY 2 – 4 (MONDAY 8 DECEMBER – WEDNESDAY 10 DECEMBER) After a good night’s sleep awaken to the sounds of traditional maskande guitar or squash-box, and enjoy a full English breakfast, which is served in the restaurant. -

North West No Fee Schools 2020
NORTH WEST NO FEE SCHOOLS 2020 NATIONAL EMIS NAME OF SCHOOL SCHOOL PHASE ADDRESS OF SCHOOL EDUCATION DISTRICT QUINTILE LEARNER NUMBER 2020 NUMBERS 2020 600100023 AMALIA PUBLIC PRIMARY PRIMARY P.O. BOX 7 AMALIA 2786 DR RUTH S MOMPATI 1 1363 600100033 ATAMELANG PRIMARY PRIMARY P.O. BOX 282 PAMPIERSTAD 8566 DR RUTH S MOMPATI 1 251 600100036 AVONDSTER PRIMARY PRIMARY P.O. BOX 335 SCHWEIZER-RENEKE 2780 DR RUTH S MOMPATI 1 171 600100040 BABUSENG PRIMARY SCHOOL PRIMARY P.O. BOX 100 LERATO 2880 NGAKA MODIRI MOLEMA 1 432 600100045 BADUMEDI SECONDARY SCHOOL SECONDARY P. O. BOX 69 RADIUM 0483 BOJANALA 1 591 600100049 BAGAMAIDI PRIMARY SCHOOL PRIMARY P.O BOX 297 HARTSWATER 8570 DR RUTH S MOMPATI 1 247 600103614 BAHENTSWE PRIMARY P.O BOX 545 DELAREYVILLE 2770 NGAKA MODIRI MOLEMA 1 119 600100053 BAISITSE PRIMARY SCHOOL PRIMARY P.O. BOX 5006 TAUNG 8584 DR RUTH S MOMPATI 1 535 600100056 BAITSHOKI HIGH SCHOOL SECONDARY PRIVATE BAG X 21 ITSOSENG 2744 NGAKA MODIRI MOLEMA 1 774 600100061 BAKGOFA PRIMARY SCHOOL PRIMARY P O BOX 1194 SUN CITY 0316 BOJANALA 1 680 600100067 BALESENG PRIMARY SCHOOL PRIMARY P. O. BOX 6 LEBOTLOANE 0411 BOJANALA 1 232 600100069 BANABAKAE PRIMARY SCHOOL PRIMARY P.O. BOX 192 LERATO 2880 NGAKA MODIRI MOLEMA 1 740 600100071 BANCHO PRIMARY SCHOOL PRIMARY PRIVATE BAG X10003 MOROKWENG 8614 DR RUTH S MOMPATI 1 60 600100073 BANOGENG MIDDLE SCHOOL SECONDARY PRIVATE BAG X 28 ITSOSENG 2744 NGAKA MODIRI MOLEMA 1 84 600100075 BAPHALANE PRIMARY SCHOOL PRIMARY P. O. BOX 108 RAMOKOKASTAD 0195 BOJANALA 1 459 600100281 BARETSE PRIMARY PRIMARY P.O. -

I METHODS of NICHE PARTITIONING BETWEEN
METHODS OF NICHE PARTITIONING BETWEEN ECUADORIAN CARNIVORES AND HABITAT PREFERENCE OF THE MARGAY ( LEOPARDUS WIEDII ) Anne-Marie C. Hodge A Thesis Submitted to the University of North Carolina Wilmington in Partial Fulfillment for the Requirements for the Degree of Master of Science Department of Biology and Marine Biology University of North Carolina Wilmington 2012 Approved By: Advisory Committee Travis Knowles Steven Emslie Marcel van Tuinen Brian Arbogast Chair Accepted by Dean, Graduate School i TABLE OF CONTENTS ABSTRACT ................................................................................................................................... iv DEDICATION .................................................................................................................................v LIST OF TABLES ......................................................................................................................... vi LIST OF FIGURES ...................................................................................................................... vii CHAPTER 1: MARGAY ACTIVITY PATTERNS AND DENSITY............................................1 Introduction ..........................................................................................................................1 Methods................................................................................................................................5 Study Location .........................................................................................................5 -

New Record for Bush Dog in Amapá State, Eastern Brazilian Amazonia
Michalski et al. Bush dogs in Eastern Brazilian Amazonia Copyright © 2015 by the IUCN/SSC Canid Specialist Group. ISSN 1478-2677 Distribution Update New record for bush dog in Amapá State, Eastern Brazilian Amazonia Lincoln J. Michalski*1,2, Tadeu G. de Oliveira3,4 and Fernanda Michalski1,2,4,5 1 Instituto Nacional de Pesquisas da Amazônia, Av. André Araújo, 2936, 69060-001 - Manaus, AM, Brazil. Email: [email protected] 2 Laboratório de Ecologia e Conservação de Vertebrados, Universidade Federal do Amapá, Rod. Juscelino Kubitscheck, km 02, 68903-419 - Macapá, AP - Brazil. 3 Departamento de Biologia, Universidade Estadual do Maranhão, Rua das Quaresmeiras, Qd-08, N°. 14, 65076-270 - São Luís, MA, Brazil. 4 Instituto Pró-Carnívoros, C.P. 10, 12940-970 - Atibaia, SP - Brazil. 5 Programa de Pós-Graduação em Biodiversidade Tropical, Universidade Federal do Amapá, Rod. Juscelino Kubitscheck, Km 02, 68903-419 - Macapá, AP - Brazil. * Correspondence author Keywords: Amapá National Forest, Amazon Forest, camera trap, geographic distribution, Speothos venaticus. Abstract Bush dogs are considered one of the lesser-known canids of South America. We report an update on their distribution in the north region of Brazil. Three bush dogs were filmed by a camera trap in Amapá Na- tional Forest, Eastern Brazilian Amazonia. The record occurred during data collection for a long-term study of medium and large vertebrates. On 28 March 2014 at 22:13h, three bush dogs passed in front of the camera. This record increases knowledge on the distribution of the species. Despite its large geographic range, bush dogs Speothos venaticus areas, bush dogs are mostly associated with well preserved areas (Lund, 1842) have been proven to be extremely difficult to locate in (Oliveira 2009) or in large forest fragments (Carretero-Pinzón 2013), the wild (DeMatteo and Loiselle 2008, DeMatteo et al. -

THE PROPOSED CONSTRUCTION of an 88Kv DISTRIBUTION POWERLINE from the EXISTING STRAATSDRIFT SUBSTATION to the PROPOSED SILWERKRAA
THE PROPOSED CONSTRUCTION OF AN 88kV DISTRIBUTION POWERLINE FROM THE EXISTING STRAATSDRIFT SUBSTATION TO THE PROPOSED SILWERKRAANS SUBSTATION WITHIN THE RAMOTSHERE MOILOA, MOSES KOTANE AND KGETLENGRIVIER LOCAL MUNICIPALITIES, NORTH WEST PROVINCE Ecological & Avifauna Component June 2017 Compiled by: Prepared for: Pachnoda Consulting CC Baagi Environmental Consultancy Lukas Niemand Pr.Sci.Nat PostNet Suite 412 PO Box 72847 Private Bag x4 Lynwood Ridge MENLO PARK Pretoria 0102 0040 Pachnoda Consulting cc Straatsdrift - Silwerkraans 88kV powerline EXECUTIVE SUMMARY Pachnoda Consulting CC was contracted by Baagi Environmental Consultancy CC to provide a terrestrial ecological report (general ecology and avifauna) for the proposed construction of an 88 kV distribution powerline from the existing Straatsdrift substation to the proposed Silwerkraans substation within the Ramotshere Moiloa, Moses Kotane and Kgetlengrivier Local Municipalities, North West Province. The project consists of two proposed corridors: Preferred Corridor (47.2 km) on the eastern section of the study area; and Alternative Corridor (45.8 km) on the western section of the study area. The terms of reference for this assessment are to: provide a general description of the affected environment concerning the avifaunal and terrestrial habitat types; conduct an assessment of all available information in order to present the following results: o typify the regional and local vegetation that will be affected by the proposed corridors; o provide an indication on the occurrence of threatened, “near- threatened”, endemic and conservation important plant, bird or animal species likely to be affected by the proposed corridors; o provide an indication of sensitive bird, fauna habitat and vegetation corresponding to the proposed corridors; o highlight areas of concern or hotspot areas; o identify potential impacts on the terrestrial ecological environment that are considered pertinent to the proposed development; o identify negative impacts and feasible mitigation options. -

Cerdocyon Thous Compiled by the Amazonian Canids Working Group – 01/2021
Literature: Cerdocyon thous Compiled by the Amazonian Canids Working Group – 01/2021 Abra FD, da Costa Canena, Garbino GST, & Medici EM. 2020. Use of unfenced highway underpasses by lowland tapirs and other medium and large mammals in central-western Brazil. Perspectives in Ecology and Conservation 18(4):247-256. Aguirre LF, Tarifa T, Wallace RB, Bernal N, Siles L, Aliaga-Rossel E & Salazar-Bravo J. 2019. Lista actualizada y comentada de los mamíferos de Bolivia. Ecología en Bolivia 54(2):107-147. Almeida, AP, Souza, TD, Marcili, A, & Labruna, MB. 2013. Novel Ehrlichia and Hepatozoon agents infecting the crab-eating fox (Cerdocyon thous) in southeastern Brazil. Journal of Medical Entomology 50(3):640-646. Aya-Cuero CA, Mosquera-Guerra F, Esquivel DA, Trujillo F, & Brooks D. 2019. Medium and large mammals of the mid Planas River basin, Colombia. Biota Colombiana 20(2):76-92. Baechli J, Albanesi S, & Bellis LM. 2021. Effectiveness of crossings as wildlife passages for mammals in the Yungas of Argentina. Journal of Nature Conservation 59:125944. https://doi.org/10.1016/j.jnc.2020.125944 . Bardeleben C, Moore RL, & Wayne RK. 2005. Isolation and Molecular Evolution of the Selenocysteine tRNA (Cf TRSP) and RNase P RNA (Cf RPPH1) Genes in the Dog Family, Canidae. Molecular Biology and Evolution 22(2):347-359. Beisiegel B de M. 1999. Contribuição ao estudo da história natural do cachorro do mato, Cerdocyon thous, e do cachorro vinagre, Speothos venaticus. PhD Dissertation. Instituto de Psicologia, Universidade de São Paulo, Brazil. Berta A. 1982. Cerdocyon thous. Mammalian Species 186(23):1-4. -

First Camera Trap Record of Bush Dogs in the State of São Paulo, Brazil
Beisiegel Bush dogs in S ão Paulo Canid News Copyright © 2009 by the IUCN/SSC Canid Specialist Group. ISSN 1478 -2677 The following is the established format for referencing this article: Beisiegel, B.M. 2009. First camera trap records of bush dogs in the state of S ão Paulo, Brazil. Canid News 12.5 [online] URL: http://www.canids.org/canidnews/12/Bush_dogs_in_Sao_Paulo.pdf. Field Report First camera trap record of bush dogs in the state of São Paulo, Brazil Beatriz M. Beisiegel Centro Nacional de Pesquisas para a Conservação dos PrePre dadores Naturais – CENAP / ICMBio. Av. dos Bandeirantes, s/n, Balneário Municipal, AtiAtibaibaia,a, CEP 12.941 -980, SP, Brazil. Email: [email protected] Keywords: Atlantic forest; camera trap ; sampling effort; Speothos venaticus . Abstract Thirteen carnivore species occur at the PECB, and a previous study using th ree camera traps (TM 500, Trail Master), over a two year period A picture of a bush dog Speothos venaticus pair obtained pictures of five of them ( crab-eating was obtained with a minimum sampling effort raccoon Procyon cancrivorus , puma Puma con- of 4,818 camera days, using seven to ten cam- color , ring-tailed coati Nasua nasua , neotropical era traps during 922 days at Parque Estadual river otter Lontra longicaudis and ocelot Leopar- Carlos Botelho, an Atlantic forest site. This dus pardalis - Beisiegel 1999). In the current picture confirms the presence of the species in study, a continuous sampling effort using the state of São Paulo, Brazil. seven to ten camera traps (Tigrinus 4.0 C, Br a- zil) begun in May 2006. -

Speothos Venaticus Across the Amazon and Other Biomes
How rare is rare? Quantifying and assessing the rarity of the bush dog Speothos venaticus across the Amazon and other biomes T ADEU G. DE O LIVEIRA,FERNANDA M ICHALSKI,ANDRÉ L. M. BOTELHO L INCOLN J. MICHALSKI,ARMANDO M. CALOURO and A RNAUD L. J. DESBIEZ Abstract The bush dog Speothos venaticus is a medium- Keywords Amazon, bush dog, camera trap, conservation, sized Neotropical canid. It is considered to be rare and its group size, rarity, relative abundance, Speothos venaticus biology and population parameters are still poorly under- To view supplementary material for this article, please visit stood. The Amazon is one of the main strongholds of this https://doi.org/./S species and is important for maintaining viable populations, as the region still holds extensive tracts of pristine habitat. We gathered field data from camera-trap studies throughout the Brazilian Amazon to estimate the relative abundance Introduction of the species and gain an understanding of its rarity, and how this compares with estimates from other vegetative for- he bush dog Speothos venaticus is a medium-sized mations and for sympatric hypercarnivores. We focused on T(c. kg) Neotropical canid that lives in packs. The spe- three pristine or partially disturbed sites and one fragmen- cies’ biology and population characteristics are poorly ted site. The estimated relative abundance of the species was understood, and it is one of the least known carnivores in – . individuals per trap-days, confirming that South America (Eisenberg & Redford, ; Zuercher ’ the species is rare. The bush dog s abundance in the et al., ; DeMatteo & Loiselle, ). -

Moses Kotane Local Municipality
Moses Kotane Local Municipality Final IDP/Budget for the Financial Year 2017/2022 4th Generation IDP Page 1 of 327 Table of Content HEADINGS PAGES SECTION A Part One 1. Mayor’s Foreword……………………………………………………………………………………. 7 – 8 2. Municipal Manager’s foreword…………………………………………………………………….. 9 – 10 Part Two 3. Introduction and Legislative Requirements…………..…………………………………….……. 12 – 14 4. Service Delivery and Budget Imlementation Plan: Progress for the last Five Years…...… 14 5. IDP/PMS/Budget Process Plan 2017/2018………………………………………………………... 15 – 16 5.1 Where are we and Where to (Vision, Mission and Values)………….……………………… 17 5.2 Municipal Vision Statement…………………………………………………………………….. 17 5.3 The Proposed Vision…………………………………………………………………………….. 17 5.4 Proposed Mission………………………………………………………………………………... 17 5.5 Municipal Values…………………………………………………………………………………. 18 5.6 Municipal Priorities…………………………………………………………………………….… 18 5.7 IDP Developmental Processes…………………………………………………………………. 19 5.8 Key Components of the IDP Processes……………………………………………………….. 20 – 29 Part Three 6. Analysis Phase 6.1 Local Orientation ………………………………………………………………………………… 30 6.2 Demographic Profile……………………………………………………………………………... 31 6.3 Racial Composition………………………………………………………………………………. 31 6.4 Ward Level Population by Age Group and Gender………………………………………….. 31 – 33 6.5 Household per ward........................................................................................................... 33 – 34 6.6 Population Distribution/composition structure and pyramid…………………………………. 32 – 33 6.7 National Mortality, -

Recueil Des Colis Postaux En Ligne SOUTH AFRICA POST OFFICE
Recueil des colis postaux en ligne ZA - South Africa SOUTH AFRICA POST OFFICE LIMITED ZAA Service de base RESDES Informations sur la réception des Oui V 1.1 dépêches (réponse à un message 1 Limite de poids maximale PREDES) (poste de destination) 1.1 Colis de surface (kg) 30 5.1.5 Prêt à commencer à transmettre des Oui données aux partenaires qui le veulent 1.2 Colis-avion (kg) 30 5.1.6 Autres données transmis 2 Dimensions maximales admises PRECON Préavis d'expédition d'un envoi Oui 2.1 Colis de surface international (poste d'origine) 2.1.1 2m x 2m x 2m Non RESCON Réponse à un message PRECON Oui (ou 3m somme de la longueur et du plus (poste de destination) grand pourtour) CARDIT Documents de transport international Oui 2.1.2 1.5m x 1.5m x 1.5m Non pour le transporteur (poste d'origine) (ou 3m somme de la longueur et du plus RESDIT Réponse à un message CARDIT (poste Oui grand pourtour) de destination) 2.1.3 1.05m x 1.05m x 1.05m Oui 6 Distribution à domicile (ou 2m somme de la longueur et du plus grand pourtour) 6.1 Première tentative de distribution Oui 2.2 Colis-avion effectuée à l'adresse physique du destinataire 2.2.1 2m x 2m x 2m Non 6.2 En cas d'échec, un avis de passage est Oui (ou 3m somme de la longueur et du plus laissé au destinataire grand pourtour) 6.3 Destinataire peut payer les taxes ou Non 2.2.2 1.5m x 1.5m x 1.5m Non droits dus et prendre physiquement (ou 3m somme de la longueur et du plus livraison de l'envoi grand pourtour) 6.4 Il y a des restrictions gouvernementales 2.2.3 1.05m x 1.05m x 1.05m Oui ou légales vous limitent dans la (ou 2m somme de la longueur et du plus prestation du service de livraison à grand pourtour) domicile.