Biomedical and Health Informatics Year in Review: Putting the Working Groups to Work Session: S22 James J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Examples of the Electronic Health Record (EHR)
Examples of the Electronic Health Record (EHR) What is Biomedical & Health Informatics? William Hersh Copyright 2020 Oregon Health & Science University Examples of the EHR • Using the Veterans Health Information Systems and Technology Architecture (VistA) • Why VistA? – State-of-the-art EHR that has transformed healthcare in the Veteran’s Health Administration (VHA) (Perlin, 2006; Byrne, 2010) – Not that pretty, but has all of the modern features of the EHR, e.g., clinical decision support (CDS), computerized provider order entry (CPOE), etc. – Distributed under open-source model, unlike most other vendors who do not even allow screen shots to be shown outside their customers’ institutions • Fascinating story (Allen, 2017), but VistA now being phased out in favor of Cerner EHR adopted by Department of Defense (VA, 2017) – https://www.ehrm.va.gov/ WhatIs5 2 1 Some details about VistA • Server written in M (formerly called MUMPS), accessed via command-line interface – Runs in commercial Intersystems Cache (on many platforms) or open-source GT.M (Linux only) • Client (called CPRS) written in Delphi and providers graphical user interface – Only runs on Windows (just about all versions) WhatIs5 3 Logging on to CPRS, the front end to VistA WhatIs5 4 2 Choosing a patient WhatIs5 5 Cover sheet – overview WhatIs5 6 3 Drilling down to details of a problem WhatIs5 7 Details of an allergy WhatIs5 8 4 Viewing vital signs over time WhatIs5 9 More details on problems WhatIs5 10 5 List of active orders WhatIs5 11 Viewing the patient’s notes WhatIs5 12 -

A Cloud Based Patient-Centered Ehealth Record
International Journal on Advances in Life Sciences, vol 7 no 1 & 2, year 2015, http://www.iariajournals.org/life_sciences/ 30 A Cloud Based Patient-Centered eHealth Record Konstantinos Koumaditis, Leonidas Katelaris, and Marinos Themistocleous Department of Digital Systems University of Piraeus Piraeus, Greece e-mail: (konkoum,lkatelaris,mthemist)@unipi.gr Abstract - This research focuses on the Patient-Centered integrated view of citizens health status. The latter is reflected e-Health (PCEH) concept by introducing its importance in EHRs and Personal Health Records (PHRs), which are and demonstrating a multidisciplinary project that being enriched and exploited by different actors and combines advanced technologies. The project links stakeholders (i.e., health and care professionals, citizens, several aspects of PCEH functionality, such as: (a) nutrition experts, hospitals, etc.) in the health ecosystem. homecare telemedicine technologies, (b) e-prescribing, e- Three general PHR models have been proposed [7]: a) the referral, e-learning and (c) state-of-the-art technologies stand-alone model, b) Electronic Health Record (EHR) like cloud computing and Service Oriented Architecture system, and c) the integrated one, which is an interoperable (SOA), will lead to an innovative integrated e-health system providing linkage with a variety of patient platform that delivers many benefits to the society, the information sources, such as EHRs, home diagnostics, economy, the industry and the research community. This insurance claims etc. The main types of health information paper provides insights of the PCEH concept and the supported by PHRs are problem lists, procedures, major current stages of the project. In doing so, we aim to illnesses, provider lists, allergy data, home-monitored data, increase the awareness of this significant work and family history, social history and lifestyle, immunizations, disseminate the knowledge gained so far through our medications and laboratory tests [8, 9]. -

Analysis of Various Techniques for Knowledge Mining in Electronic Health Records
European Journal of Molecular & Clinical Medicine ISSN 2515-8260 Volume 7, Issue 11, 2020 Analysis of various techniques for Knowledge Mining in Electronic Health Records 1V.Deepa, 2P.Mohamed Fathimal 1Research Scholar , 2 Assistant Professor ,SRM Institute of Science & Technology, Vadapalani ,Chennai Abstract With the fast development of digital communication all over the world ,each domain including healthcare sector has led to new dimension . “Health Informatics,” are the term used to coin application of IT for better healthcare services. Its applications helps to maintain the health record of individuals, in digital form known as the Electronic Health Record .This paper reviews the Electronic health records (EHR) in healthcare organizations. These digital records can help to support clinical activities that have the ability to improve quality of health and to reduce the costs. The aim of this paper is to study the detail analysis of the structure and the components of electronic health record.This paper reviews different techniques for handling the information in the text and different components of the electronic health record include daily charting, physical assessment, medication, discharge history, physical examination and test procedures. Keywords: Physical Assessment, Survey medication, Semantics, patience, Information Quality, Health care policy planning I INTRODUCTION In healthcare domain, more than 1100 electronic health record vendors are available all over the world. National policy and guidelines have been framed for EHR dealers and medical benefits. EHR collects real time medical data which stores patient’s information in a digital format like a history chart. As all the information are accessible in a single file, extraction of medical data from EMRs (electronic medical records) for analysis is very effective for the analysis. -

Free/Libre Open Source Software (FLOSS) in Healthcare: Showcase of OSCAR, a Canadian Project
Free/Libre Open Source Software (FLOSS) in Healthcare: Showcase of OSCAR, A Canadian Project David Chan, MD, CCFP, MSc, FCFP Associate Professor Department of Family Medicine McMaster University, Hamilton, Ontario, Canada 1 QNUCM Nov 12, 2006 11/12/06 PPresentationresentation outlineoutline What is Free/Libre Open Source Software (FLOSS) What is OSCAR A Quick Tour Question & Answer 2 QNUCM Nov 12, 2006 11/12/06 ResourcesResources http://oscarmcmaster.org (please join) The Cathedral and the Bazaar (Eric Raymond) Open Sources: Voices from the Open Source Revolution (O’Reilly publisher) The Success of Open Source (Steven Weber) The Future of Ideas: the Fate of the Commons in a Connected World (Lawrence Lessig) http://en.wikipedia.org/wiki/Open_source 3 QNUCM Nov 12, 2006 11/12/06 4 QNUCM Nov 12, 2006 11/12/06 5 QNUCM Nov 12, 2006 11/12/06 6 QNUCM Nov 12, 2006 11/12/06 7 QNUCM Nov 12, 2006 11/12/06 WWhhaatt isis F FLLOOSSSS?? Source code versus machine code 8 QNUCM Nov 12, 2006 11/12/06 9 QNUCM Nov 12, 2006 11/12/06 10 QNUCM Nov 12, 2006 11/12/06 WWhhaatt isis F FLLOOSSSS?? Source code versus machine code Free as in “libre” not “gratis” (“freedom” not “free beer”) – freedom from single vendor lock-in – freedom from proprietary “non-standards” – freedom from DEATH (average life-span of most vendors)! – freedom to adapt to CHANGE: health care environment (e.g. Primary Care Reform) technology (e.g. hardware, software, and the Internet) standards (e.g. Infoway 2009: HL7, LOINC, SNOWMED-CT) new knowledge (e.g. -
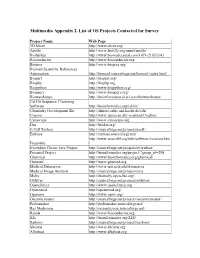
PDF File, 212 KB
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -
Step-By-Step Install Guide OSCAR Mcmaster Ehealth CMS on Ubuntu
Global Open Versity eHealth Labs Install Guide OSCAR McMaster CMS on Linux Ubuntu 10.04 LTS v1.5 Global Open Versity e-Health Management Systems Hands-on Labs Training Manual Step-by-Step Install Guide OSCAR McMaster CMS on Linux Ubuntu 10.04 LTS (Lucid Lynx) Kefa Rabah Global Open Versity, Vancouver Canada [email protected] www.globalopenversity.org Table of Contents Page No. STEP-BY-STEP INSTALL GUIDE OSCAR MCMASTER CMS ON LINUX UBUNTU 10.04 LTS 3 1.0 Introduction 3 Part 1: Install & Configure Linux Ubuntu 10.04 LTS Desktop 4 Step 1: Getting Started & Hardware Pre-requisites 4 Step 2: Install Linux Ubuntu 10.04 LTS 4 Step 3: Update Ubuntu 10.04 Operating Systems 4 Part 2: Installing OSCAR McMaster CMS on Linux Ubuntu 8.04 LTS Server 5 Step 1: Preamble 5 Part 3: Installing the Infrastructure Packages 6 Step 1: Install OpenSSH Package 6 Step 2: Install Sun-Java6-JDK Package 7 Step 3: Install Ant Package 8 Step 4: Install MySQL Database Server Package 8 Step 5: Install Apache Tomcat 6 Package 9 Step 6: Install Pretty Good Privacy (PGP) Package 9 Step 7: Install CVS 10 Step 8: Install PostgreSQL database server 10 Step 9: Install UNZIP Package 10 Step 10: Install Network Time Protocol (NTP) Service 11 Step 11: Reboot the server 11 Step 12: Configuring the Base Packages 11 Step 13: Configuring Tomcat6 12 Step 14: Secure Apache Tomcat6 using SSL 13 Part 5: Install OSCAR CMS Server 15 Step 1: Download & Install OSCAR 15 Step 2: Create OSCAR Database on MySQL 16 Part 6: Enabling New eCharts 18 Step 1: Enable eChart 18 Part 7: Drugref2/3 -

Comparison of Open-Source Electronic Health Record Systems Based on Functional and User Performance Criteria
Original Article Healthc Inform Res. 2019 April;25(2):89-98. https://doi.org/10.4258/hir.2019.25.2.89 pISSN 2093-3681 • eISSN 2093-369X Comparison of Open-Source Electronic Health Record Systems Based on Functional and User Performance Criteria Saptarshi Purkayastha1, Roshini Allam1, Pallavi Maity1, Judy W. Gichoya2 1Department of BioHealth Informatics, Indiana University–Purdue University Indianapolis, Indianapolis, IN, USA 2Dotter Institute, Oregon Health and Science University, Portland, OR, USA Objectives: Open-source Electronic Health Record (EHR) systems have gained importance. The main aim of our research is to guide organizational choice by comparing the features, functionality, and user-facing system performance of the five most popular open-source EHR systems. Methods: We performed qualitative content analysis with a directed approach on recently published literature (2012–2017) to develop an integrated set of criteria to compare the EHR systems. The functional criteria are an integration of the literature, meaningful use criteria, and the Institute of Medicine’s functional requirements of EHR, whereas the user-facing system performance is based on the time required to perform basic tasks within the EHR system. Results: Based on the Alexa web ranking and Google Trends, the five most popular EHR systems at the time of our study were OSHERA VistA, GNU Health, the Open Medical Record System (OpenMRS), Open Electronic Medical Record (OpenEMR), and OpenEHR. We also found the trends in popularity of the EHR systems and the locations where they were more popular than others. OpenEMR met all the 32 functional criteria, OSHERA VistA met 28, OpenMRS met 12 fully and 11 partially, OpenEHR-based EHR met 10 fully and 3 partially, and GNU Health met the least with only 10 criteria fully and 2 partially. -
Step-By-Step Install Guide OSCAR Mcmaster CMS on Linux Ubuntu 10.04 LTS (Lucid Lynx)
Global Open Versity eHealth Labs Install Guide OSCAR McMaster CMS on Linux Ubuntu 10.04 LTS v1.5 Global Open Versity e-Health Management Systems Hands-on Labs Training Manual Step-by-Step Install Guide OSCAR McMaster CMS on Linux Ubuntu 10.04 LTS (Lucid Lynx) Kefa Rabah Global Open Versity, Vancouver Canada [email protected] www.globalopenversity.org Table of Contents Page No. STEP-BY-STEP INSTALL GUIDE OSCAR MCMASTER CMS ON LINUX UBUNTU 10.04 LTS 3 1.0 Introduction 3 Part 1: Install & Configure Linux Ubuntu 10.04 LTS Desktop 4 Step 1: Getting Started & Hardware Pre-requisites 4 Step 2: Install Linux Ubuntu 10.04 LTS 4 Step 3: Update Ubuntu 10.04 Operating Systems 4 Part 2: Installing OSCAR McMaster CMS on Linux Ubuntu 8.04 LTS Server 5 Step 1: Preamble 5 Part 3: Installing the Infrastructure Packages 6 Step 1: Install OpenSSH Package 6 Step 2: Install Sun-Java6-JDK Package 7 Step 3: Install Ant Package 8 Step 4: Install MySQL Database Server Package 8 Step 5: Install Apache Tomcat 6 Package 9 Step 6: Install Pretty Good Privacy (PGP) Package 9 Step 7: Install CVS 10 Step 8: Install PostgreSQL database server 10 Step 9: Install UNZIP Package 10 Step 10: Install Network Time Protocol (NTP) Service 11 Step 11: Reboot the server 11 Step 12: Configuring the Base Packages 11 Step 13: Configuring Tomcat6 12 Step 14: Secure Apache Tomcat6 using SSL 13 Part 5: Install OSCAR CMS Server 15 Step 1: Download & Install OSCAR 15 Step 2: Create OSCAR Database on MySQL 16 Part 6: Enabling New eCharts 18 Step 1: Enable eChart 18 Part 7: Drugref2/3 -

Advances in Information Technology 0.Pdf
ADVANCES IN INFORMATION TECHNOLOGY AND COMMUNICATION IN HEALTH Studies in Health Technology and Informatics This book series was started in 1990 to promote research conducted under the auspices of the EC programmes’ Advanced Informatics in Medicine (AIM) and Biomedical and Health Research (BHR) bioengineering branch. A driving aspect of international health informatics is that telecommunication technology, rehabilitative technology, intelligent home technology and many other components are moving together and form one integrated world of information and communication media. The complete series has been accepted in Medline. Volumes from 2005 onwards are available online. Series Editors: Dr. O. Bodenreider, Dr. J.P. Christensen, Prof. G. de Moor, Prof. A. Famili, Dr. U. Fors, Prof. A. Hasman, Prof. E.J.S. Hovenga, Prof. L. Hunter, Dr. I. Iakovidis, Dr. Z. Kolitsi, Mr. O. Le Dour, Dr. A. Lymberis, Prof. J. Mantas, Prof. M.A. Musen, Prof. P.F. Niederer, Prof. A. Pedotti, Prof. O. Rienhoff, Prof. F.H. Roger France, Dr. N. Rossing, Prof. N. Saranummi, Dr. E.R. Siegel and Dr. P. Wilson Volume 143 Recently published in this series Vol. 142. J.D. Westwood, S.W. Westwood, R.S. Haluck, H.M. Hoffman, G.T. Mogel, R. Phillips, R.A. Robb and K.G. Vosburgh (Eds.), Medicine Meets Virtual Reality 17 – NextMed: Design for/the Well Being Vol. 141. E. De Clercq et al. (Eds.), Collaborative Patient Centred eHealth – Proceedings of the HIT@HealthCare 2008 joint event: 25th MIC Congress, 3rd International Congress Sixi, Special ISV-NVKVV Event, 8th Belgian eHealth Symposium Vol. 140. P.H. Dangerfield (Ed.), Research into Spinal Deformities 6 Vol. -

Open Source Et Gestion De L'information Médicale : Aperçu Des Projets Existants
Open source et gestion de l'information médicale : aperçu des projets existants Ateliers CISP Club 2011 – Alexandre BRULET Open Source : bref historique... 1969 : UNIX (Bell Labs°) 1975 : distribué à des universités pour « fins éducatives » avec les codes sources... 1977 : projet BSD 1984 : projet GNU (R. Stallman) (sources partagées) 1985 : Free Software Fondation (cadre légal – licence GPL) 1989 : licence BSD modifiée (open source) 1991 : noyau GNU/Linux ? OS dérivés (NetBSD, FreeBSD, SunOS ...) 1993 : Slackware 1993 : Debian 1994 : Red Hat > 50 OS dérivés > 100 OS dérivés 1998 : MPL (ex : SUSE) > 50 OS dérivés (Knopix, Ubuntu...) (Mandriva, Fedora ...) 1999 : licence Apache UNESCO 2004 : logiciels libres patrimoine de l'humanité (…) > 35 licences libres recensées sur wikipédia (PHP, Cecil, MIT, CPL, W3C, etc.) Open source : un fonctionnement communautaire ● La « pyramide » Linux : développeurs / 'maintainers' / chefs de projet sys USB net (...) L. Torvalds / A. Morton ● Système de «patchs» : publics, signés, discutés, soumis, (approuvés) ● Versions stables régulières et archivées (mirroirs) ● Système de «paquets» permettant la cohérence des OS ● Mode de fonctionnement repris par la plupart des distributions basées sur Linux ainsi que leurs « filles » : Debian → Ubuntu, Slackware → Zenwalk, RedHat → Fedora, etc. ● Idem pour les logiciels (xfce/gnome/kde, OOo, Gimp, Firefox, etc.) Le monde open source : un immense agrégat de communautés... OS OS OS Projets GNU OS OS OS Noyau OS OS OS Noyau OS OS Projets BSD Programme open source Programme propriétaire Système OS d'exploitation Quid des logiciels médicaux ? Petit tour du monde de l'open source médical à partir d'une liste proposée par Wikipédia. 1. Logiciels médicaux francophones : MedinTux ● Petite communauté depuis 2005 (marseille), licence CeCiLL ● DMI cabinet / hôpital - objectif = ergonomie ● Programmes serveurs et clients, consultation web possible. -

Pipenightdreams Osgcal-Doc Mumudvb Mpg123-Alsa Tbb
pipenightdreams osgcal-doc mumudvb mpg123-alsa tbb-examples libgammu4-dbg gcc-4.1-doc snort-rules-default davical cutmp3 libevolution5.0-cil aspell-am python-gobject-doc openoffice.org-l10n-mn libc6-xen xserver-xorg trophy-data t38modem pioneers-console libnb-platform10-java libgtkglext1-ruby libboost-wave1.39-dev drgenius bfbtester libchromexvmcpro1 isdnutils-xtools ubuntuone-client openoffice.org2-math openoffice.org-l10n-lt lsb-cxx-ia32 kdeartwork-emoticons-kde4 wmpuzzle trafshow python-plplot lx-gdb link-monitor-applet libscm-dev liblog-agent-logger-perl libccrtp-doc libclass-throwable-perl kde-i18n-csb jack-jconv hamradio-menus coinor-libvol-doc msx-emulator bitbake nabi language-pack-gnome-zh libpaperg popularity-contest xracer-tools xfont-nexus opendrim-lmp-baseserver libvorbisfile-ruby liblinebreak-doc libgfcui-2.0-0c2a-dbg libblacs-mpi-dev dict-freedict-spa-eng blender-ogrexml aspell-da x11-apps openoffice.org-l10n-lv openoffice.org-l10n-nl pnmtopng libodbcinstq1 libhsqldb-java-doc libmono-addins-gui0.2-cil sg3-utils linux-backports-modules-alsa-2.6.31-19-generic yorick-yeti-gsl python-pymssql plasma-widget-cpuload mcpp gpsim-lcd cl-csv libhtml-clean-perl asterisk-dbg apt-dater-dbg libgnome-mag1-dev language-pack-gnome-yo python-crypto svn-autoreleasedeb sugar-terminal-activity mii-diag maria-doc libplexus-component-api-java-doc libhugs-hgl-bundled libchipcard-libgwenhywfar47-plugins libghc6-random-dev freefem3d ezmlm cakephp-scripts aspell-ar ara-byte not+sparc openoffice.org-l10n-nn linux-backports-modules-karmic-generic-pae -

Adopting an Open Source Hospital Information System to Manage Healthcare Institutions
LIFE: International Journal of Health and Life-Sciences ISSN 2454-5872 Bouidi et al., 2017 Volume 3 Issue 3, pp. 38-57 Date of Publication: 18th December 2017 DOI-https://dx.doi.org/10.20319/lijhls.2017.33.3857 This paper can be cited as: Bouidi, Y, Azzouzi Idrissi, M & Rais, N. (2017). Adopting an Open Source Hospital Information System to Manage Healthcare Institutions. LIFE: International Journal of Health and Life-Sciences, 3(3), 38-57. This work is licensed under the Creative Commons Attribution-Non Commercial 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc/4.0/ or send a letter to Creative Commons, PO Box 1866, Mountain View, CA 94042, USA. ADOPTING AN OPEN SOURCE HOSPITAL INFORMATION SYSTEM TO MANAGE HEALTHCARE INSTITUTIONS Youssef Bouidi Laboratory of Informatics, Modeling and Systems (LIMS), University of Sidi Mohamed Ben Abdallah (USMBA), Fez, Morocco [email protected] Mostafa Azzouzi Idrissi Laboratory of Informatics, Modeling and Systems (LIMS), University of Sidi Mohamed Ben Abdallah (USMBA), Fez, Morocco [email protected] Noureddine Rais Laboratory of Informatics, Modeling and Systems (LIMS), University of Sidi Mohamed Ben Abdallah (USMBA), Fez, Morocco [email protected] Abstract Our paper is a comparative study of different Open Source Hospital Information Systems (OSHISs). We chose open source because of problems in healthcare management like budget, resources and computerization. A literature review did not allow us to find a similar comparison, which explains the great interest of our study. Firstly, we retrieved nine OSHISs: MediBoard, OpenEMR, OpenMRS, OpenHospital, HospitalOS, PatientOS, Care2x, MedinTux and HOSxP.