Lepidoptera: Bombycoidea)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Entomologica 31 1997 Entomologica18/05/98
View metadata, citation and similar papers at core.ac.uk Entomologica, Bari, 31,brought (1997): to 191-195 you by CORE provided by Università degli Studi di Bari: Open Journal Systems RENATO SPICCIARELLI Dipartimento di Biologia, Difesa e Biotecnologie Agro-Forestali Università della Basilicata, Potenza Primi reperti di larve di Acanthobrahmaea europaea (Hrtg.) (Lepidoptera: Brahmaeidae) su Phillyrea latifolia L.* ABSTRACT REPORTS OF LARVAE OF ACANTHOBRAHMAEA EUROPAEA (HARTIG) ON PHILLYREA LATIFOLIA L. In typical environment of Acanthobrahmaea europaea (Vulture Mt., Basilicata, Southern Italy) several beatings were carried out from April to May 1997 on plants of ash, privet and phillyrea, to search for the presence of larvae of this species. Each plant was examined either by direct obser- vation or by shacking branches over a cloth. At middle of May, a group of six larvae of the second instar was found on a plant of Phillyrea latifolia L. This report does confirm a previous report on the same plant species in an area very distant from the Vulture Mt. (Campomaggiore, Potenza prov.). Because the larvae were found out of the “Riserva Naturale Orientata Statale Grotticelle”, a new delimitation of this protected area seems suitable. Key words: Lepidoptera, Acanthobrahmaea europaea, Phillyrea latifolia, food plant. INTRODUZIONE In questi ultimi anni, la Acanthobrahmaea europaea (HARTIG, 1963; 1966) è stata reperita più volte anche in aree della Basilicata diverse dal Vulture, con cacce al lume negli scali ferroviari e lungo le rive dei fiumi Ofanto e Basento (PARENZAN, 1977; BERTACCINI et al., 1995), nonché in Campania, ad Aquilonia- Scalo (PARENZAN, l.c.), località prossima al confine con la Basilicata. -

Moths Light a Way? by John Pickering, Tori Staples and Rebecca Walcott
SOUTHERN LEPIDOPTERISTS NEWS VOLUME 38 NO4. (2016), PG. 331 SAVE ALL SPECIES – MOTHS LIGHT A WAY? BY JOHN PICKERING, TORI STAPLES AND REBECCA WALCOTT Abstract -- What would it take to save all species from snakes, and stinging insects, they pose no health risk. extinction? A new initiative, Save all species, plans to Moths are an exceedingly species-rich group, for which answer this question and provide the tools we need to do the diversity at a terrestrial site will typically exceed any so by 2050. Here we consider the merits and problems other taxon except for beetles. Because moth larvae are associated with inventorying moths to help decide which restricted in their diet to specific host taxa, differences in terrestrial areas to protect. We compare the the assemblages of resident moth species could reflect scientifically-described moth fauna of the British Isles differences across sites in plants and other hosts. If which, with 2,441 species, is taxonomically complete, that’s true, we may be able to use moth inventories as with 11,806 described species from North America north efficient proxies to compare surrounding plant of Mexico, the fauna of which is not fully described. As communities. a percentage of the described moth fauna, there are fewer “macro” moths (Geometroidea, Drepanoidea, Inventorying moths presents challenges, notably, Noctuoidea, Bombycoidea, Lasiocampidae) in the sampling smaller species, describing thousands of British Isles (34.9%) than those known for the United species new to science, and identifying specimens States and Canada (46.1%). We present data on 1,254 accurately. Our experience is that we can identify 99% species for an intensively-studied site in Clarke County, of moths from digital images to species, species-groups, Georgia and consider whether species in the British Isles which contain species of similar appearance, or are generally smaller than ones in Georgia. -

Modular Structure, Sequence Diversification and Appropriate
www.nature.com/scientificreports OPEN Modular structure, sequence diversifcation and appropriate nomenclature of seroins produced Received: 17 July 2018 Accepted: 14 February 2019 in the silk glands of Lepidoptera Published: xx xx xxxx Lucie Kucerova1, Michal Zurovec 1,2, Barbara Kludkiewicz1, Miluse Hradilova3, Hynek Strnad3 & Frantisek Sehnal1,2 Seroins are small lepidopteran silk proteins known to possess antimicrobial activities. Several seroin paralogs and isoforms were identifed in studied lepidopteran species and their classifcation required detailed phylogenetic analysis based on complete and verifed cDNA sequences. We sequenced silk gland-specifc cDNA libraries from ten species and identifed 52 novel seroin cDNAs. The results of this targeted research, combined with data retrieved from available databases, form a dataset representing the major clades of Lepidoptera. The analysis of deduced seroin proteins distinguished three seroin classes (sn1-sn3), which are composed of modules: A (includes the signal peptide), B (rich in charged amino acids) and C (highly variable linker containing proline). The similarities within and between the classes were 31–50% and 22.5–25%, respectively. All species express one, and in exceptional cases two, genes per class, and alternative splicing further enhances seroin diversity. Seroins occur in long versions with the full set of modules (AB1C1B2C2B3) and/or in short versions that lack parts or the entire B and C modules. The classes and the modular structure of seroins probably evolved prior to the split between Trichoptera and Lepidoptera. The diversity of seroins is refected in proposed nomenclature. Te silk spun by caterpillars is a composite material based on two protein agglomerates that have been known for centuries as fbroin and sericin. -

Lepidoptera of North America 5
Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera by Valerio Albu, 1411 E. Sweetbriar Drive Fresno, CA 93720 and Eric Metzler, 1241 Kildale Square North Columbus, OH 43229 April 30, 2004 Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Cover illustration: Blueberry Sphinx (Paonias astylus (Drury)], an eastern endemic. Photo by Valeriu Albu. ISBN 1084-8819 This publication and others in the series may be ordered from the C.P. Gillette Museum of Arthropod Diversity, Department of Bioagricultural Sciences and Pest Management Colorado State University, Fort Collins, CO 80523 Abstract A list of 1531 species ofLepidoptera is presented, collected over 15 years (1988 to 2002), in eleven southern West Virginia counties. A variety of collecting methods was used, including netting, light attracting, light trapping and pheromone trapping. The specimens were identified by the currently available pictorial sources and determination keys. Many were also sent to specialists for confirmation or identification. The majority of the data was from Kanawha County, reflecting the area of more intensive sampling effort by the senior author. This imbalance of data between Kanawha County and other counties should even out with further sampling of the area. Key Words: Appalachian Mountains, -

Butterflies and Moths of Gwinnett County, Georgia, United States
Heliothis ononis Flax Bollworm Moth Coptotriche aenea Blackberry Leafminer Argyresthia canadensis Apyrrothrix araxes Dull Firetip Phocides pigmalion Mangrove Skipper Phocides belus Belus Skipper Phocides palemon Guava Skipper Phocides urania Urania skipper Proteides mercurius Mercurial Skipper Epargyreus zestos Zestos Skipper Epargyreus clarus Silver-spotted Skipper Epargyreus spanna Hispaniolan Silverdrop Epargyreus exadeus Broken Silverdrop Polygonus leo Hammock Skipper Polygonus savigny Manuel's Skipper Chioides albofasciatus White-striped Longtail Chioides zilpa Zilpa Longtail Chioides ixion Hispaniolan Longtail Aguna asander Gold-spotted Aguna Aguna claxon Emerald Aguna Aguna metophis Tailed Aguna Typhedanus undulatus Mottled Longtail Typhedanus ampyx Gold-tufted Skipper Polythrix octomaculata Eight-spotted Longtail Polythrix mexicanus Mexican Longtail Polythrix asine Asine Longtail Polythrix caunus (Herrich-Schäffer, 1869) Zestusa dorus Short-tailed Skipper Codatractus carlos Carlos' Mottled-Skipper Codatractus alcaeus White-crescent Longtail Codatractus yucatanus Yucatan Mottled-Skipper Codatractus arizonensis Arizona Skipper Codatractus valeriana Valeriana Skipper Urbanus proteus Long-tailed Skipper Urbanus viterboana Bluish Longtail Urbanus belli Double-striped Longtail Urbanus pronus Pronus Longtail Urbanus esmeraldus Esmeralda Longtail Urbanus evona Turquoise Longtail Urbanus dorantes Dorantes Longtail Urbanus teleus Teleus Longtail Urbanus tanna Tanna Longtail Urbanus simplicius Plain Longtail Urbanus procne Brown Longtail -

Lepidoptera Sphingidae:) of the Caatinga of Northeast Brazil: a Case Study in the State of Rio Grande Do Norte
212212 JOURNAL OF THE LEPIDOPTERISTS’ SOCIETY Journal of the Lepidopterists’ Society 59(4), 2005, 212–218 THE HIGHLY SEASONAL HAWKMOTH FAUNA (LEPIDOPTERA SPHINGIDAE:) OF THE CAATINGA OF NORTHEAST BRAZIL: A CASE STUDY IN THE STATE OF RIO GRANDE DO NORTE JOSÉ ARAÚJO DUARTE JÚNIOR Programa de Pós-Graduação em Ciências Biológicas, Departamento de Sistemática e Ecologia, Universidade Federal da Paraíba, 58059-900, João Pessoa, Paraíba, Brasil. E-mail: [email protected] AND CLEMENS SCHLINDWEIN Departamento de Botânica, Universidade Federal de Pernambuco, Av. Prof. Moraes Rego, s/n, Cidade Universitária, 50670-901, Recife, Pernambuco, Brasil. E-mail:[email protected] ABSTRACT: The caatinga, a thorn-shrub succulent savannah, is located in Northeastern Brazil and characterized by a short and irregular rainy season and a severe dry season. Insects are only abundant during the rainy months, displaying a strong seasonal pat- tern. Here we present data from a yearlong Sphingidae survey undertaken in the reserve Estação Ecológica do Seridó, located in the state of Rio Grande do Norte. Hawkmoths were collected once a month during two subsequent new moon nights, between 18.00h and 05.00h, attracted with a 160-watt mercury vapor light. A total of 593 specimens belonging to 20 species and 14 genera were col- lected. Neogene dynaeus, Callionima grisescens, and Hyles euphorbiarum were the most abundant species, together comprising up to 82.2% of the total number of specimens collected. These frequent species are residents of the caatinga of Rio Grande do Norte. The rare Sphingidae in this study, Pseudosphinx tetrio, Isognathus australis, and Cocytius antaeus, are migratory species for the caatinga. -

Wildlife Review Cover Image: Hedgehog by Keith Kirk
Dumfries & Galloway Wildlife Review Cover Image: Hedgehog by Keith Kirk. Keith is a former Dumfries & Galloway Council ranger and now helps to run Nocturnal Wildlife Tours based in Castle Douglas. The tours use a specially prepared night tours vehicle, complete with external mounted thermal camera and internal viewing screens. Each participant also has their own state- of-the-art thermal imaging device to use for the duration of the tour. This allows participants to detect animals as small as rabbits at up to 300 metres away or get close enough to see Badgers and Roe Deer going about their nightly routine without them knowing you’re there. For further information visit www.wildlifetours.co.uk email [email protected] or telephone 07483 131791 Contributing photographers p2 Small White butterfly © Ian Findlay, p4 Colvend coast ©Mark Pollitt, p5 Bittersweet © northeastwildlife.co.uk, Wildflower grassland ©Mark Pollitt, p6 Oblong Woodsia planting © National Trust for Scotland, Oblong Woodsia © Chris Miles, p8 Birdwatching © castigatio/Shutterstock, p9 Hedgehog in grass © northeastwildlife.co.uk, Hedgehog in leaves © Mark Bridger/Shutterstock, Hedgehog dropping © northeastwildlife.co.uk, p10 Cetacean watch at Mull of Galloway © DGERC, p11 Common Carder Bee © Bob Fitzsimmons, p12 Black Grouse confrontation © Sergey Uryadnikov/Shutterstock, p13 Black Grouse male ©Sergey Uryadnikov/Shutterstock, Female Black Grouse in flight © northeastwildlife.co.uk, Common Pipistrelle bat © Steven Farhall/ Shutterstock, p14 White Ermine © Mark Pollitt, -

Redalyc.A Review of the Genus Ectropa Wallengren, 1863 with Descriptions of a New Genus and Six New Species (Lepidoptera: Chryso
SHILAP Revista de Lepidopterología ISSN: 0300-5267 [email protected] Sociedad Hispano-Luso-Americana de Lepidopterología España Kurshakov, P. A.; Zolotuhin, V. V. A review of the genus Ectropa Wallengren, 1863 with descriptions of a new genus and six new species (Lepidoptera: Chrysopolomidae) SHILAP Revista de Lepidopterología, vol. 41, núm. 164, octubre-diciembre, 2013, pp. 431-447 Sociedad Hispano-Luso-Americana de Lepidopterología Madrid, España Available in: http://www.redalyc.org/articulo.oa?id=45530406003 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative 431-447 A review of the genus 2/12/13 16:37 Página 431 SHILAP Revta. lepid., 41 (164), diciembre 2013: 431-447 eISSN: 2340-4078 ISSN: 0300-5267 A review of the genus Ectropa Wallengren, 1863 with descriptions of a new genus and six new species (Lepidoptera: Chrysopolomidae) P. A. Kurshakov & V. V. Zolotuhin Summary The genus Ectropa Wallengren, 1863, is revised, and a new species, E. adam Kurshakov & Zolotuhin, sp. n., is described. Based on the wing pattern and male genitalia characters, a new genus Ectropona Kurshakov & Zolotuhin, gen. n., related to Ectropa, is erected here, with Ectropona dargei Kurshakov & Zolotuhin, sp. n., as a type-species. Five more new species are also described: Ectropona dargei Kurshakov & Zolotuhin, sp. n., E. larsa Kurshakov & Zolotuhin, sp. n., E. kubwa Kurshakov & Zolotuhin, sp. n., E. aarviki Kurshakov & Zolotuhin, sp. n., and E. revelli Kurshakov & Zolotuhin, sp. -

National Parks Association of the Australian Capital Territory Inc
Volume 53 Number 2 June 2016 National Parks Association of the Australian Capital Territory Inc. Burning Aranda Bushland Canberra Nature Map Jagungal Wilderness NPA Bulletin Volume 53 number 2 June 2016 Articles by contributors may not necessarily reflect association opinion or objectives. CONTENTS NPA outings program, June – September 2016 ...............13–16 From the Committee ................................................................2 Bushwalks Rod Griffiths and Christine Goonrey Exciting Rendezvous Valley pack walk ..........................17 The vital work of the National Parks Australia Council ..........3 Esther Gallant Rod Griffiths Mount Tantangara ...........................................................18 NPA's Nature Play program .....................................................3 Brian Slee Graham Scully Pretty Plain ......................................................................19 Aranda Bushland's recent hazard-reduction burn ....................4 Brian Slee Judy Kelly, with Michael Doherty and John Brickhill Glenburn Precinct news..........................................................20 Obituaries .................................................................................6 Col McAlister Book reviews. Leaf Litter, exploring the Mysteries................21 The National Rock Garden ......................................................7 of a Hidden World by Rachel Tonkin Compiled by Kevin McCue Judy Kelly Stolen .......................................................................................7 -

The Mcguire Center for Lepidoptera and Biodiversity
Supplemental Information All specimens used within this study are housed in: the McGuire Center for Lepidoptera and Biodiversity (MGCL) at the Florida Museum of Natural History, Gainesville, USA (FLMNH); the University of Maryland, College Park, USA (UMD); the Muséum national d’Histoire naturelle in Paris, France (MNHN); and the Australian National Insect Collection in Canberra, Australia (ANIC). Methods DNA extraction protocol of dried museum specimens (detailed instructions) Prior to tissue sampling, dried (pinned or papered) specimens were assigned MGCL barcodes, photographed, and their labels digitized. Abdomens were then removed using sterile forceps, cleaned with 100% ethanol between each sample, and the remaining specimens were returned to their respective trays within the MGCL collections. Abdomens were placed in 1.5 mL microcentrifuge tubes with the apex of the abdomen in the conical end of the tube. For larger abdomens, 5 mL microcentrifuge tubes or larger were utilized. A solution of proteinase K (Qiagen Cat #19133) and genomic lysis buffer (OmniPrep Genomic DNA Extraction Kit) in a 1:50 ratio was added to each abdomen containing tube, sufficient to cover the abdomen (typically either 300 µL or 500 µL) - similar to the concept used in Hundsdoerfer & Kitching (1). Ratios of 1:10 and 1:25 were utilized for low quality or rare specimens. Low quality specimens were defined as having little visible tissue inside of the abdomen, mold/fungi growth, or smell of bacterial decay. Samples were incubated overnight (12-18 hours) in a dry air oven at 56°C. Importantly, we also adjusted the ratio depending on the tissue type, i.e., increasing the ratio for particularly large or egg-containing abdomens. -
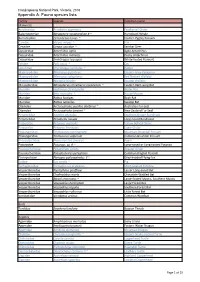
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -

Australian Sphingidae – DNA Barcodes Challenge Current Species Boundaries and Distributions
Australian Sphingidae – DNA Barcodes Challenge Current Species Boundaries and Distributions Rodolphe Rougerie1*¤, Ian J. Kitching2, Jean Haxaire3, Scott E. Miller4, Axel Hausmann5, Paul D. N. Hebert1 1 University of Guelph, Biodiversity Institute of Ontario, Guelph, Ontario, Canada, 2 Natural History Museum, Department of Life Sciences, London, United Kingdom, 3 Honorary Attache´, Muse´um National d’Histoire Naturelle de Paris, Le Roc, Laplume, France, 4 National Museum of Natural History, Smithsonian Institution, Washington, DC, United States of America, 5 Bavarian State Collection of Zoology, Section Lepidoptera, Munich, Germany Abstract Main Objective: We examine the extent of taxonomic and biogeographical uncertainty in a well-studied group of Australian Lepidoptera, the hawkmoths (Sphingidae). Methods: We analysed the diversity of Australian sphingids through the comparative analysis of their DNA barcodes, supplemented by morphological re-examinations and sequence information from a nuclear marker in selected cases. The results from the analysis of Australian sphingids were placed in a broader context by including conspecifics and closely related taxa from outside Australia to test taxonomic boundaries. Results: Our results led to the discovery of six new species in Australia, one case of erroneously synonymized species, and three cases of synonymy. As a result, we establish the occurrence of 75 species of hawkmoths on the continent. The analysis of records from outside Australia also challenges the validity of current taxonomic boundaries in as many as 18 species, including Agrius convolvuli (Linnaeus, 1758), a common species that has gained adoption as a model system. Our work has revealed a higher level of endemism than previously recognized. Most (90%) Australian sphingids are endemic to the continent (45%) or to Australia, the Pacific Islands and the Papuan and Wallacean regions (45%).