Population Dynamics of Invasive Argentine Ant
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Winged Ants (Hymenoptera: Formicidae) Presence in Twigs on the Leaf Litter of Atlantic Forest
Biota Neotropica 19(3): e20180694, 2019 www.scielo.br/bn ISSN 1676-0611 (online edition) Inventory Winged ants (Hymenoptera: Formicidae) presence in twigs on the leaf litter of Atlantic Forest Tae Tanaami Fernandes1 , Rogério R. Silva2, Débora Rodrigues de Souza-Campana2, Otávio Guilherme Morais da Silva2 & Maria Santina de Castro Morini1* 1Universidade de Mogi das Cruzes, Laboratório de Mirmecologia do Alto Tietê, Rua Dr. Cândido Xavier de Almeida e Souza, 200, CEP 08780-911, Mogi das Cruzes, SP, Brasil 2Museu Paraense Emílio Goeldi, Coordenação de Ciências da Terra e Ecologia, Avenida Perimetral, 1901, Terra Firme, CEP 66077-830, Belém, PA, Brasil *Corresponding author: Maria Santina de Castro Morini, e-mail: [email protected] FERNANDES, T. T., SILVA, R. R., SOUZA-CAMPANA, D. R., SILVA, O. G. M., MORINI, M. S. C. Winged ants (Hymenoptera: Formicidae) presence in twigs on the leaf litter of Atlantic Forest. Biota Neotropica 19(3): e20180694. http://dx.doi.org/10.1590/1676-0611-BN-2018-0694 Abstract: In the leaf litter, ants have various nesting resources available, such as live or dead trunks, twigs, leaves, fruits and seeds. On the twigs, there are adults and immature individuals, but also the queen and winged. The production of wings requires time and energy from the colony. The objective of this study was to investigate the presence of winged in ant colonies in twigs on the leaf litter. Our prediction is that the richness and abundance of winged in twigs are the greatest in rainy months. We collected all twigs with ants in 552 plots with 16 m2, totaling 8,832 m2 of leaf litter, in areas located in the Brazilian Atlantic Domain. -

Ants in French Polynesia and the Pacific: Species Distributions and Conservation Concerns
Ants in French Polynesia and the Pacific: species distributions and conservation concerns Paul Krushelnycky Dept of Plant and Environmental Protection Sciences, University of Hawaii, Honolulu, Hawaii Hervé Jourdan Centre de Biologie et de Gestion des Populations, INRA/IRD, Nouméa, New Caledonia The importance of ants • In most ecosystems, form a substantial portion of a communities’ biomass (1/3 of animal biomass and ¾ of insect biomass in Amazon rainforest) Photos © Alex Wild The importance of ants • In most ecosystems, form a substantial portion of a communities’ biomass (1/3 of animal biomass and ¾ of insect biomass in Amazon rainforest) • Involved in many important ecosystem processes: predator/prey relationships herbivory seed dispersal soil turning mutualisms Photos © Alex Wild The importance of ants • Important in shaping evolution of biotic communities and ecosystems Photos © Alex Wild Ants in the Pacific • Pacific archipelagoes the most remote in the world • Implications for understanding ant biogeography (patterns of dispersal, species/area relationships, community assembly) • Evolution of faunas with depauperate ant communities • Consequent effects of ant introductions Hypoponera zwaluwenburgi Ants in the Amblyopone zwaluwenburgi Pacific – current picture Ponera bableti Indigenous ants in the Pacific? Approx. 30 - 37 species have been labeled “wide-ranging Pacific natives”: Adelomyrmex hirsutus Ponera incerta Anochetus graeffei Ponera loi Camponotus chloroticus Ponera swezeyi Camponotus navigator Ponera tenuis Camponotus rufifrons -

ORIGINAL ARTICLE a Novel Intracellular Mutualistic Bacterium in the Invasive Ant Cardiocondyla Obscurior
The ISME Journal (2016) 10, 376–388 © 2016 International Society for Microbial Ecology All rights reserved 1751-7362/16 www.nature.com/ismej ORIGINAL ARTICLE A novel intracellular mutualistic bacterium in the invasive ant Cardiocondyla obscurior Antonia Klein1,8, Lukas Schrader1,8, Rosario Gil2, Alejandro Manzano-Marín2, Laura Flórez3, David Wheeler5, John H Werren6, Amparo Latorre2,7, Jürgen Heinze1, Martin Kaltenpoth3,4, Andrés Moya2 and Jan Oettler1 1Institut für Zoologie, Universität Regensburg, Regensburg, Germany; 2Institut Canvanilles de Biodiversitat i Biologia Evolutiva (ICBiBE), Parc Cientific de la Universitat de Valencia, Paterna (Valencia), Spain; 3Max Planck Institute for Chemical Ecology, Jena, Germany; 4Johannes Gutenberg University Mainz, Institute for Zoology, Department for Evolutionary Ecology, Mainz, Germany; 5Institute of Fundamental Sciences, Massey University, Palmerston North, New Zealand; 6Department of Biology, University Rochester, Rochester, NY, USA and 7Área de Genómica y Salud de la Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana (FISABIO)–Salud Pública, Valencia, Spain The evolution of eukaryotic organisms is often strongly influenced by microbial symbionts that confer novel traits to their hosts. Here we describe the intracellular Enterobacteriaceae symbiont of theinvasiveantCardiocondyla obscurior, ‘Candidatus Westeberhardia cardiocondylae’. Upon metamorphosis, Westeberhardia is found in gut-associated bacteriomes that deteriorate following eclosion. Only queens maintain Westeberhardia in the ovarian nurse cells from where the symbionts are transmitted to late-stage oocytes during nurse cell depletion. Functional analyses of the streamlined genome of Westeberhardia (533 kb, 23.41% GC content) indicate that neither vitamins nor essential amino acids are provided for the host. However, the genome encodes for an almost complete shikimate pathway leading to 4-hydroxyphenylpyruvate, which could be converted into tyrosine by the host. -

Taxonomy and Distribution of the Argentine Ant, Linepithema Humile (Hymenoptera: Formicidae)
SYSTEMATICS Taxonomy and Distribution of the Argentine Ant, Linepithema humile (Hymenoptera: Formicidae) ALEXANDER L. WILD Department of Entomology, University of California at Davis, Davis, CA 95616 Ann. Entomol. Soc. Am. 97(6): 1204Ð1215 (2004) ABSTRACT The taxonomy of an invasive pest species, the Argentine ant, is reviewed. Linepithema humile (Mayr) 1868 is conÞrmed as the valid name for the Argentine ant. Morphological variation and species boundaries of L. humile are examined, with emphasis on populations from the antÕs native range in southern South America. Diagnoses and illustrations are provided for male, queen, and worker castes. Collection records of L. humile in South America support the idea of a native distribution closely associated with major waterways in lowland areas of the Parana´ River drainage, with recent intro- ductions into parts of Argentina, Brazil, Chile, Colombia, Ecuador, and Peru. KEY WORDS Argentine ant, Linepithema humile, taxonomy, invasive species THE ARGENTINE ANT, Linepithema humile (Mayr) 1868, MCSN, MCZC, MHNG, MZSP, NHMB, NHMW, and is among the worldÕs most successful invasive species. USNM; see below for explanation of abbreviations). This native South American insect has become a cos- Taxonomic confusion over L. humile extends be- mopolitan pest, particularly in the Mediterranean cli- yond museum collections. At least one important mates of North America, Chile, South Africa, Austra- study, seeking to explain Argentine ant population lia, and southern Europe (Suarez et al. 2001). regulation in the native range through phorid para- Argentine ants have been implicated in the decline of sitism (Orr and Seike 1998), initially targeted the native arthropod (Cole et al. 1992) and vertebrate wrong Linepithema species (Orr et al. -

Notes on Ants (Hymenoptera: Formicidae) from Gambia (Western Africa)
ANNALS OF THE UPPER SILESIAN MUSEUM IN BYTOM ENTOMOLOGY Vol. 26 (online 010): 1–13 ISSN 0867-1966, eISSN 2544-039X (online) Bytom, 08.05.2018 LECH BOROWIEC1, SEBASTIAN SALATA2 Notes on ants (Hymenoptera: Formicidae) from Gambia (Western Africa) http://doi.org/10.5281/zenodo.1243767 1 Department of Biodiversity and Evolutionary Taxonomy, University of Wrocław, Przybyszewskiego 65, 51-148 Wrocław, Poland e-mail: [email protected], [email protected] Abstract: A list of 35 ant species or morphospecies collected in Gambia is presented, 9 of them are recorded for the first time from the country:Camponotus cf. vividus, Crematogaster cf. aegyptiaca, Dorylus nigricans burmeisteri SHUCKARD, 1840, Lepisiota canescens (EMERY, 1897), Monomorium cf. opacum, Monomorium cf. salomonis, Nylanderia jaegerskioeldi (MAYR, 1904), Technomyrmex pallipes (SMITH, 1876), and Trichomyrmex abyssinicus (FOREL, 1894). A checklist of 82 ant species recorded from Gambia is given. Key words: ants, faunistics, Gambia, new country records. INTRODUCTION Ants fauna of Gambia (West Africa) is poorly known. Literature data, AntWeb and other Internet resources recorded only 59 species from this country. For comparison from Senegal, which surrounds three sides of Gambia, 89 species have been recorded so far. Both of these records seem poor when compared with 654 species known from the whole western Africa (SHUCKARD 1840, ANDRÉ 1889, EMERY 1892, MENOZZI 1926, SANTSCHI 1939, LUSH 2007, ANTWIKI 2017, ANTWEB 2017, DIAMÉ et al. 2017, TAYLOR 2018). Most records from Gambia come from general web checklists of species. Unfortunately, they lack locality data, date of sampling, collector name, coordinates of the locality and notes on habitats. -

Floral Volatiles Play a Key Role in Specialized Ant Pollination Clara De Vega
FLORAL VOLATILES PLAY A KEY ROLE IN SPECIALIZED ANT POLLINATION CLARA DE VEGA1*, CARLOS M. HERRERA1, AND STEFAN DÖTTERL2,3 1 Estación Biológica de Doñana, Consejo Superior de Investigaciones Científicas (CSIC), Avenida de Américo Vespucio s/n, 41092 Sevilla, Spain 2 University of Bayreuth, Department of Plant Systematics, 95440 Bayreuth, Germany 3 Present address: University of Salzburg, Organismic Biology, Hellbrunnerstr. 34, 5020 Salzburg, Austria Running title —Floral scent and ant pollination * For correspondence. E-mail [email protected] Tel: +34 954466700 Fax: + 34 954621125 1 ABSTRACT Chemical signals emitted by plants are crucial to understanding the ecology and evolution of plant-animal interactions. Scent is an important component of floral phenotype and represents a decisive communication channel between plants and floral visitors. Floral 5 volatiles promote attraction of mutualistic pollinators and, in some cases, serve to prevent flower visitation by antagonists such as ants. Despite ant visits to flowers have been suggested to be detrimental to plant fitness, in recent years there has been a growing recognition of the positive role of ants in pollination. Nevertheless, the question of whether floral volatiles mediate mutualisms between ants and ant-pollinated plants still remains largely unexplored. 10 Here we review the documented cases of ant pollination and investigate the chemical composition of the floral scent in the ant-pollinated plant Cytinus hypocistis. By using chemical-electrophysiological analyses and field behavioural assays, we examine the importance of olfactory cues for ants, identify compounds that stimulate antennal responses, and evaluate whether these compounds elicit behavioural responses. Our findings reveal that 15 floral scent plays a crucial role in this mutualistic ant-flower interaction, and that only ant species that provide pollination services and not others occurring in the habitat are efficiently attracted by floral volatiles. -

Terrestrial Arthropod Surveys on Pagan Island, Northern Marianas
Terrestrial Arthropod Surveys on Pagan Island, Northern Marianas Neal L. Evenhuis, Lucius G. Eldredge, Keith T. Arakaki, Darcy Oishi, Janis N. Garcia & William P. Haines Pacific Biological Survey, Bishop Museum, Honolulu, Hawaii 96817 Final Report November 2010 Prepared for: U.S. Fish and Wildlife Service, Pacific Islands Fish & Wildlife Office Honolulu, Hawaii Evenhuis et al. — Pagan Island Arthropod Survey 2 BISHOP MUSEUM The State Museum of Natural and Cultural History 1525 Bernice Street Honolulu, Hawai’i 96817–2704, USA Copyright© 2010 Bishop Museum All Rights Reserved Printed in the United States of America Contribution No. 2010-015 to the Pacific Biological Survey Evenhuis et al. — Pagan Island Arthropod Survey 3 TABLE OF CONTENTS Executive Summary ......................................................................................................... 5 Background ..................................................................................................................... 7 General History .............................................................................................................. 10 Previous Expeditions to Pagan Surveying Terrestrial Arthropods ................................ 12 Current Survey and List of Collecting Sites .................................................................. 18 Sampling Methods ......................................................................................................... 25 Survey Results .............................................................................................................. -

Can a Ten-Year Old Decision Not to Eradicate Be Re-Visited?
The distribution of Argentine ant in New Zealand: Can a ten-year old decision not to eradicate be re-visited? J.G. Charles1, D.M. Suckling2, D.J. Allan1, K.J. Froud1, P.R. Dentener1, P.G. Connolly1 and H. Verberne3 1HortResearch, Private Bag 92 169, Auckland, New Zealand 2HortResearch, P.O. Box 51, Lincoln, New Zealand 3AgriQuality, Private Bag 3080, Hamilton, New Zealand Corresponding author: [email protected] Summary Decisions to eradicate, contain or accept new invaders are made in the face of often complex interactions between biological, social, economic and environmental factors. The outcomes of this dynamic mix may change over time, so that, for any species, a decision made in the past may differ to that made in the present or future. This is illustrated by the Argentine ant, Linepithema humile, which established in New Zealand in about 1990, with no attempt at eradication or containment. Changing attitudes and control options since then have led to a study of the geographical distribution of the ant throughout New Zealand over a 10 week period from mid-March until the end of May 2001. Foraging workers were collected in jam-baited traps over a 24 h period. Traps were deployed in ‘starburst arrays’ of up to 33 traps, or in groups of four, or singly at selected locations and habitats where L. humile was, or might be expected to be, a problem. Argentine ants were caught in 84 traps (out of 2216) and were shown to be widely, though patchily, distributed throughout New Zealand, especially in the north of the North Island. -

Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects
insects Review (Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects Kayli R. Sieber 1 , Taylor Dorman 1, Nicholas Newell 1 and Hua Yan 1,2,* 1 Department of Biology, University of Florida, Gainesville, FL 32611, USA; kayli.sieber@ufl.edu (K.R.S.); taylor.dorman@ufl.edu (T.D.); nicholas.newell@ufl.edu (N.N.) 2 Center for Smell and Taste, University of Florida, Gainesville, FL 32611, USA * Correspondence: hua.yan@ufl.edu; Tel.: +1-352-273-4983 Simple Summary: Social insects, namely ants, bees, and termites, are among the most numerous and successful animals on Earth. This is due to a variety of features: highly cooperative behavior performed by colony members and their specialization on a variety of tasks. Diverse physiological and behavioral specializations are regulated not only by the genetic system, but also by the epige- netic system which alters gene expressions without modifying the genetic code. This review will summarize recent advancements in such studies in eusocial insects. Abstract: Eusocial insects, such as bees, ants, and wasps of the Hymenoptera and termites of the Blattodea, are able to generate remarkable diversity in morphology and behavior despite being genetically uniform within a colony. Most eusocial insect species display caste structures in which reproductive ability is possessed by a single or a few queens while all other colony members act Citation: Sieber, K.R.; Dorman, T.; as workers. However, in some species, caste structure is somewhat plastic, and individuals may Newell, N.; Yan, H. (Epi)Genetic switch from one caste or behavioral phenotype to another in response to certain environmental cues. -
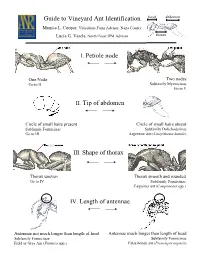
I. Petiole Node II. Tip of Abdomen IV. Length of Antennae Guide to Vineyard Ant Identification III. Shape of Thorax
Guide to Vineyard Ant Identification head abdomen Monica L. Cooper, Viticulture Farm Advisor, Napa County Lucia G. Varela, North Coast IPM Advisor thorax I. Petiole node One Node Two nodes Go to II Subfamily Myrmicinae Go to V II. Tip of abdomen Circle of small hairs present Circle of small hairs absent Subfamily Formicinae Subfamily Dolichoderinae Go to III Argentine Ant (Linepithema humile) III. Shape of thorax Thorax uneven Thorax smooth and rounded Go to IV Subfamily Formicinae Carpenter ant (Camponotus spp.) IV. Length of antennae Antennae not much longer than length of head Antennae much longer than length of head Subfamily Formicinae Subfamily Formicinae Field or Gray Ant (Formica spp.) False honey ant (Prenolepis imparis) head abdomen Petiole with two nodes Subfamily Myrmicinae (V-VIII) thorax V. Dorsal side of Thorax & Antennae One pair of spines on thorax No spines on thorax 12 segmented antennae 10 segmented antennae Go to VI Solenopsis molesta and Solenopsis xyloni VI. Underside of head No brush of bristles Brush of long bristles Go to VII Harvester ants (Pogonomyrmex californicus and P. brevispinosis) VII. Head and Thorax With hairs Without hairs Go to VIII Cardiocondyla mauritanica VIII. Head and Thorax With many parallel furrows Without parallel furrows Profile of thorax rounded Profile of thorax not evenly rounded Pavement ant (Tetramorium “species E”) Pheidole californica Argentine Ant (Linepithema humile), subfamily Dolichoderinae Exotic species 3-4 mm in length Deep brown to light black Move rapidly in distinct trails Feed on honeydew Shallow nests (2 inches from soil surface) Alex Wild Does not bite or sting Carpenter Ant (Camponotus spp.), subfamily Formicinae Large ant: >6 mm in length Dark color with smooth, rounded thorax Workers most active at dusk and night One of most abundant and widespread genera worldwide Generalist scavengers and predators: feed on dead and living insects, nectar, fruit juices and Jack K. -

A Second Species of Lepisiota Spreading Across the Canary
Fragmenta entomologica, 50 (1): 61-64 (2018) eISSN: 2284-4880 (online version) pISSN: 0429-288X (print version) Short scientific note Submitted: February 28th, 2018 - Accepted: May 24th, 2018 - Published: June 29th, 2018 Yet another alien: a second species of Lepisiota spreading across the Canary Islands, Spain (Hymenoptera: Formicidae) Enrico SCHIFANI 1,*, Vincenzo GENTILE 2, Antonio SCUPOLA 3, Xavier ESPADALER 4 1 Section Animal Biology, Department STEBICEF, University of Palermo - Via Archirafi 18, I-90123 Palermo, Italy [email protected] 2 C.so Umberto I 301, 80058 Torre Annunziata (NA), Italy - [email protected] 3 Museo civico di Storia Naturale di Verona - Lungadige Porta Vittoria 9, 37129 Verona, Italy - [email protected] 4 CREAF, Universitat Autònoma de Barcelona - 08193 Cerdanyola del Vallés, Spain - [email protected] * Corresponding author Abstract The Canary Islands are a biologically important archipelago hosting many unique species, whose myrmecofauna is peculiarly rich in both endemic and introduced species. Lepisiota frauenfeldi cfr. kantarensis Forel, 1911 is reported for the first time from Fuerteventura and Tenerife. It is the second species of Lepisiota introduced in the archipelago in the last few years, and one of the few documented cas- es in which Lepisiota frauenfeldi (Mayr, 1855) s.l. acts as a successful tramp species. Comments are also given on taxonomic problems involving the L. frauenfeldi-group and related taxa. Finally, new additional information and comments are presented on the distribution of other alien ants species from the Canary Islands [Lasius neglectus Van Loon, Boomsma & Andrásfalvy, 1990, Lepisiota capensis (Mayr, 1862) and Paratrechina longicornis (Latreille, 1802)]. Key words: Macaronesia, Lepisiota frauenfeldi ssp. -

Hymenoptera: Formicidae: Dolichoderinae) from Colombia
Revista Colombiana de Entomología 37 (1): 159-161 (2011) 159 Scientific note The first record of the genus Gracilidris (Hymenoptera: Formicidae: Dolichoderinae) from Colombia Primer registro del género Gracilidris (Hymenoptera: Formicidae: Dolichoderinae) para Colombia ROBERTO J. GUERRERO1 and CATALINA SANABRIA2 Abstract: The dolichoderine ant genus Gracilidris and its sole species, G. pombero, are recorded for the first time for Colombia from populations from the foothills of the Colombian Amazon basin. Comments and hypotheses about the biogeography of the genus are discussed. Key words: Ants. Biodiversity. Caquetá. Colombian Amazon. Grazing systems. Resumen: El género dolicoderino de hormigas Gracilidris y su única especie, G. pombero, son registrados por primera vez para Colombia, de poblaciones provenientes del piedemonte de la cuenca Amazónica colombiana. Algunos comen- tarios e hipótesis sobre la biogeografía del género son discutidos. Palabras clave: Hormigas. Biodiversidad. Caquetá. Amazonas colombiano. Pasturas ganaderas. Introduction distribution of the genus Gracilidris in South America. We also discuss each of the dolichoderine genera that occur in Currently, dolichoderine ants (Hymenoptera: Formicidae: Colombia. Dolichoderinae) include 28 extant genera (Bolton et al. 2006; Fisher 2009) distributed in four tribes according to Methods the latest higher level classification of the ant subfamily Dolichoderinae (Ward et al. 2010). Eleven of those extant We separated G. pombero specimens from all ants collected genera occur in the New World: Bothriomyrmex, Dolicho- in the project “Amaz_BD: Biodiversidad de los paisajes derus, Liometopum, Tapinoma, and Technomyrmex have a Amazónicos, determinantes socio-económicos y produc- global distribution, while Anillidris, Azteca, Dorymyrmex, ción de bienes y servicios”. This research was carried out in Forelius, Gracilidris, and Linepithema are endemic to the Caquetá department located in the western foothills of the New World.