A Multi-Level Investigation of the Genetic Relationship Between
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Redefining the Specificity of Phosphoinositide-Binding by Human
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.20.163253; this version posted June 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins Nilmani Singh1†, Adriana Reyes-Ordoñez1†, Michael A. Compagnone1, Jesus F. Moreno Castillo1, Benjamin J. Leslie2, Taekjip Ha2,3,4,5, Jie Chen1* 1Department of Cell & Developmental Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801; 2Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205; 3Department of Biophysics, Johns Hopkins University, Baltimore, MD 21218; 4Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21205; 5Howard Hughes Medical Institute, Baltimore, MD 21205, USA †These authors contributed equally to this work. *Correspondence: [email protected]. bioRxiv preprint doi: https://doi.org/10.1101/2020.06.20.163253; this version posted June 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. ABSTRACT Pleckstrin homology (PH) domains are presumed to bind phosphoinositides (PIPs), but specific interaction with and regulation by PIPs for most PH domain-containing proteins are unclear. Here we employed a single-molecule pulldown assay to study interactions of lipid vesicles with full-length proteins in mammalian whole cell lysates. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

The Proteomic Landscape of Resting and Activated CD4+ T Cells Reveal Insights Into Cell Differentiation and Function
International Journal of Molecular Sciences Article The Proteomic Landscape of Resting and Activated CD4+ T Cells Reveal Insights into Cell Differentiation and Function Yashwanth Subbannayya 1 , Markus Haug 1, Sneha M. Pinto 1, Varshasnata Mohanty 2, Hany Zakaria Meås 1, Trude Helen Flo 1, T.S. Keshava Prasad 2 and Richard K. Kandasamy 1,* 1 Centre of Molecular Inflammation Research (CEMIR), Department of Clinical and Molecular Medicine (IKOM), Norwegian University of Science and Technology, 7491 Trondheim, Norway; [email protected] (Y.S.); [email protected] (M.H.); [email protected] (S.M.P.); [email protected] (H.Z.M.); trude.fl[email protected] (T.H.F.) 2 Center for Systems Biology and Molecular Medicine, Yenepoya (Deemed to be University), Mangalore 575018, India; [email protected] (V.M.); [email protected] (T.S.K.P.) * Correspondence: [email protected] Abstract: CD4+ T cells (T helper cells) are cytokine-producing adaptive immune cells that activate or regulate the responses of various immune cells. The activation and functional status of CD4+ T cells is important for adequate responses to pathogen infections but has also been associated with auto-immune disorders and survival in several cancers. In the current study, we carried out a label-free high-resolution FTMS-based proteomic profiling of resting and T cell receptor-activated (72 h) primary human CD4+ T cells from peripheral blood of healthy donors as well as SUP-T1 cells. We identified 5237 proteins, of which significant alterations in the levels of 1119 proteins were observed between resting and activated CD4+ T cells. -

Immune Adaptor SKAP1 Acts a Scaffold for Polo-Like Kinase 1 (PLK1)
www.nature.com/scientificreports OPEN Immune adaptor SKAP1 acts a scafold for Polo-like kinase 1 (PLK1) for the optimal cell cycling Received: 14 September 2018 Accepted: 6 June 2019 of T-cells Published: xx xx xxxx Monika Raab1,2, Klaus Strebhardt1 & Christopher E. Rudd2,3,4 While the immune cell adaptor protein SKAP1 mediates LFA-1 activation induced by antigen-receptor (TCR/CD3) ligation on T-cells, it is unclear whether the adaptor interacts with other mediators of T-cell function. In this context, the serine/threonine kinase, polo-like kinase (PLK1) regulates multiple steps in the mitotic and cell cycle progression of mammalian cells. Here, we show that SKAP1 is phosphorylated by and binds to PLK1 for the optimal cycling of T-cells. PLK1 binds to the N-terminal residue serine 31 (S31) of SKAP1 and the interaction is needed for optimal PLK1 kinase activity. Further, siRNA knock- down of SKAP1 reduced the rate of T-cell division concurrent with a delay in the expression of PLK1, Cyclin A and pH3. Reconstitution of these KD cells with WT SKAP1, but not the SKAP1 S31 mutant, restored normal cell division. SKAP1-PLK1 binding is dynamically regulated during the cell cycle of T-cells. Our fndings identify a novel role for SKAP1 in the regulation of PLK1 and optimal cell cycling needed for T-cell clonal expansion in response to antigenic activation. Polo-like kinase 1 (PLK1) is a serine/threonine kinase that regulates the mitosis of mammalian cells. It is a mem- ber of the Polo-like serine/threonine kinase (PLKs) family that are structurally conserved from yeast to mam- mals. -

Quantitative Interactomics in Primary T Cells Unveils TCR Signal
Quantitative interactomics in primary T cells unveils TCR signal diversification extent and dynamics Guillaume Voisinne, Kristof Kersse, Karima Chaoui, Liaoxun Lu, Julie Chaix, Lichen Zhang, Marisa Goncalves Menoita, Laura Girard, Youcef Ounoughene, Hui Wang, et al. To cite this version: Guillaume Voisinne, Kristof Kersse, Karima Chaoui, Liaoxun Lu, Julie Chaix, et al.. Quantitative interactomics in primary T cells unveils TCR signal diversification extent and dynamics. Nature Immunology, Nature Publishing Group, 2019, 20 (11), pp.1530 - 1541. 10.1038/s41590-019-0489-8. hal-03013469 HAL Id: hal-03013469 https://hal.archives-ouvertes.fr/hal-03013469 Submitted on 23 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. RESOURCE https://doi.org/10.1038/s41590-019-0489-8 Quantitative interactomics in primary T cells unveils TCR signal diversification extent and dynamics Guillaume Voisinne 1, Kristof Kersse1, Karima Chaoui2, Liaoxun Lu3,4, Julie Chaix1, Lichen Zhang3, Marisa Goncalves Menoita1, Laura Girard1,5, Youcef Ounoughene1, Hui Wang3, Odile Burlet-Schiltz2, Hervé Luche5,6, Frédéric Fiore5, Marie Malissen1,5,6, Anne Gonzalez de Peredo 2, Yinming Liang 3,6*, Romain Roncagalli 1* and Bernard Malissen 1,5,6* The activation of T cells by the T cell antigen receptor (TCR) results in the formation of signaling protein complexes (signalo somes), the composition of which has not been analyzed at a systems level. -

A Graph-Theoretic Approach to Model Genomic Data and Identify Biological Modules Asscociated with Cancer Outcomes
A Graph-Theoretic Approach to Model Genomic Data and Identify Biological Modules Asscociated with Cancer Outcomes Deanna Petrochilos A dissertation presented in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2013 Reading Committee: Neil Abernethy, Chair John Gennari, Ali Shojaie Program Authorized to Offer Degree: Biomedical Informatics and Health Education UMI Number: 3588836 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. UMI 3588836 Published by ProQuest LLC (2013). Copyright in the Dissertation held by the Author. Microform Edition © ProQuest LLC. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, MI 48106 - 1346 ©Copyright 2013 Deanna Petrochilos University of Washington Abstract Using Graph-Based Methods to Integrate and Analyze Cancer Genomic Data Deanna Petrochilos Chair of the Supervisory Committee: Assistant Professor Neil Abernethy Biomedical Informatics and Health Education Studies of the genetic basis of complex disease present statistical and methodological challenges in the discovery of reliable and high-confidence genes that reveal biological phenomena underlying the etiology of disease or gene signatures prognostic of disease outcomes. This dissertation examines the capacity of graph-theoretical methods to model and analyze genomic information and thus facilitate using prior knowledge to create a more discrete and functionally relevant feature space. -

Systematic Elucidation of Neuron-Astrocyte Interaction in Models of Amyotrophic Lateral Sclerosis Using Multi-Modal Integrated Bioinformatics Workflow
ARTICLE https://doi.org/10.1038/s41467-020-19177-y OPEN Systematic elucidation of neuron-astrocyte interaction in models of amyotrophic lateral sclerosis using multi-modal integrated bioinformatics workflow Vartika Mishra et al.# 1234567890():,; Cell-to-cell communications are critical determinants of pathophysiological phenotypes, but methodologies for their systematic elucidation are lacking. Herein, we propose an approach for the Systematic Elucidation and Assessment of Regulatory Cell-to-cell Interaction Net- works (SEARCHIN) to identify ligand-mediated interactions between distinct cellular com- partments. To test this approach, we selected a model of amyotrophic lateral sclerosis (ALS), in which astrocytes expressing mutant superoxide dismutase-1 (mutSOD1) kill wild-type motor neurons (MNs) by an unknown mechanism. Our integrative analysis that combines proteomics and regulatory network analysis infers the interaction between astrocyte-released amyloid precursor protein (APP) and death receptor-6 (DR6) on MNs as the top predicted ligand-receptor pair. The inferred deleterious role of APP and DR6 is confirmed in vitro in models of ALS. Moreover, the DR6 knockdown in MNs of transgenic mutSOD1 mice attenuates the ALS-like phenotype. Our results support the usefulness of integrative, systems biology approach to gain insights into complex neurobiological disease processes as in ALS and posit that the proposed methodology is not restricted to this biological context and could be used in a variety of other non-cell-autonomous communication -

Inflammatory, Regulatory, and Autophagy Co-Expression Modules
Durocher et al. Journal of Neuroinflammation (2019) 16:56 https://doi.org/10.1186/s12974-019-1433-4 RESEARCH Open Access Inflammatory, regulatory, and autophagy co-expression modules and hub genes underlie the peripheral immune response to human intracerebral hemorrhage Marc Durocher1, Bradley P. Ander1, Glen Jickling1, Farah Hamade1, Heather Hull1, Bodie Knepp1, Da Zhi Liu1, Xinhua Zhan1, Anh Tran1, Xiyuan Cheng1, Kwan Ng1, Alan Yee1, Frank R. Sharp1 and Boryana Stamova1,2* Abstract Background: Intracerebral hemorrhage (ICH) has a high morbidity and mortality. The peripheral immune system and cross-talk between peripheral blood and brain have been implicated in the ICH immune response. Thus, we delineated the gene networks associated with human ICH in the peripheral blood transcriptome. We also compared the differentially expressed genes in blood following ICH to a prior human study of perihematomal brain tissue. Methods: We performed peripheral blood whole-transcriptome analysis of ICH and matched vascular risk factor control subjects (n = 66). Gene co-expression network analysis identified groups of co-expressed genes (modules) associated with ICH and their most interconnected genes (hubs). Mixed-effects regression identified differentially expressed genes in ICH compared to controls. Results: Of seven ICH-associated modules, six were enriched with cell-specific genes: one neutrophil module, one neutrophil plus monocyte module, one T cell module, one Natural Killer cell module, and two erythroblast modules. The neutrophil/monocyte modules were enriched in inflammatory/immune pathways; the T cell module in T cell receptor signaling genes; and the Natural Killer cell module in genes regulating alternative splicing, epigenetic, and post-translational modifications. -

Genetic Commonalities in Gynecologic Cancers Using Publicly Available Genome-Wide Association Study Summary Results: an Exploratory Meta-Analysis
Title Page Genetic Commonalities in Gynecologic Cancers Using Publicly Available Genome-Wide Association Study Summary Results: An Exploratory Meta-Analysis by Mark Francis Vater BS Biology, Northern Kentucky University, 2014 Submitted to the Graduate Faculty of the Graduate School of Public Health in partial fulfillment of the requirements for the degree of Master of Science University of Pittsburgh 2021 Committee Membership Page UNIVERSITY OF PITTSBURGH GRADUATE SCHOOL OF PUBLIC HEALTH This thesis was presented by Mark Francis Vater It was defended on August 4, 2021 and approved by Jeanine Buchanich, Associate Professor, Department of Biostatistics Ada Youk, Associate Professor, Department of Biostatistics John Shaffer, Assistant Professor, Department of Human Genetics Thesis Advisor: Jenna Carlson, Assistant Professor, Department of Biostatistics ii Copyright © by Mark Vater 2021 iii Abstract Genetic Commonalities in Gynecologic Cancers Using Publicly Available Genome-Wide Association Study Summary Results: An Exploratory Meta-Analysis Mark Francis Vater, MS University of Pittsburgh, 2021 Abstract Public Health Significance: Gynecologic cancers are responsible for millions of deaths worldwide every year. The goal of this research is to further scientific understanding of such cancers, potentially leading to improved diagnosis and treatment. The public health significance of this project is to uncover potential genetic underpinnings of gynecologic cancers, aiding in efforts to reduce the mortality rate of gynecologic cancers, which is imperative in protecting the health of susceptible persons across the globe. Gynecologic cancers are those which arise in the female reproductive system, chiefly, those of the ovaries, cervix, vulva, endometrium, and vagina. These conditions present a serious threat to the health of susceptible persons, leading to nearly three million deaths worldwide each year. -
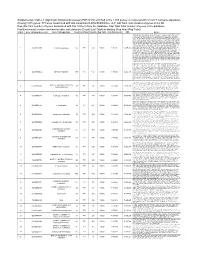
(FDR<0.05) Enriched in the 1015 Genes Co-Clustered with Known T
Supplementary Table 3. Significant biological processes (FDR<0.05) enriched in the 1,015 genes co-clustered with known T cell gene signatures. Among 1,015 genes, 771 were associated with GO annotation in DAVID database v6.7. List Total: total number of genes in my list. Pop Hits: total number of genes associated with this GO term from the database. Pop Total: total number of genes in the database. Fold Enrichment: relative enrichment ratio, calculated by (Count)/(List Total) divided by (Pop Hits)/(Pop Total). Index Gene Ontology Accession Gene Ontology Name Count List Total Pop Hits Pop Total Fold Enrichment FDR Genes AQP9, C1QC, B2M, LILRA1, LILRA2, CLEC4E, S1PR4, LILRA4, IFNG, LILRA6, CLEC4A, VNN1, ERAP2, FAS, CRTAM, C5AR1, GBP5, NCF2, NCF1, NCF4, SERPING1, HLA-DQA2, HLA-DQA1, PDCD1LG2, LILRB1, CCR9, C1QA, C1QB, LILRB2, CCR7, CCR6, UNC13D, CCR5, CD40LG, CCR4, LILRB3, CCR2, LILRB4, HLA-DPA1, VSIG4, HLA-DRA, IL1R2, IL1R1, HLA-DRB1, OAS3, ACP5, OAS1, OAS2, CD74, IFI35, ZAP70, FCER1G, HLA-DRB5, HLA-DPB1, HLA-DOA, HLA-DOB, DHX58, BLNK, IL23R, KIR2DS4, CD300C, SLAMF7, OASL, RGS1, APOL1, CD300A, HMHB1, CD209, CLEC7A, LY86, LY9, CLNK, FCRL4, SH2D1A, NOD2, HAMP, CCL3L1, CCL3L3, TICAM2, ICAM1, GZMA, CMKLR1, LY96, WAS, IL18BP, LAX1, TNFSF12- TNFSF13, HLA-DQB1, CSF2, GPR183, CCR1, GPR65, CXCL9, NCF1C, IL7R, CLEC10A, CCL24, CCL22, CYP27B1, CCL23, FCGR1C, FTHL3, FCGR1A, FCGR1B, BCL3, C2, CD27, CD28, FYB, IL18R1, IL7, CD1C, CTLA4, CCL19, CD1B, CD1A, TRIM22, CD180, CD1E, CCL18, CCL17, CCL13, FCGR2B, FCGR2C, P2RY14, LIME1, CD14, IL16, IL18, TLR1, TNFSF15, -

Agricultural University of Athens
ΓΕΩΠΟΝΙΚΟ ΠΑΝΕΠΙΣΤΗΜΙΟ ΑΘΗΝΩΝ ΣΧΟΛΗ ΕΠΙΣΤΗΜΩΝ ΤΩΝ ΖΩΩΝ ΤΜΗΜΑ ΕΠΙΣΤΗΜΗΣ ΖΩΙΚΗΣ ΠΑΡΑΓΩΓΗΣ ΕΡΓΑΣΤΗΡΙΟ ΓΕΝΙΚΗΣ ΚΑΙ ΕΙΔΙΚΗΣ ΖΩΟΤΕΧΝΙΑΣ ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ ΑΘΗΝΑ 2020 ΔΙΔΑΚΤΟΡΙΚΗ ΔΙΑΤΡΙΒΗ Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Genome-wide association analysis and gene network analysis for (re)production traits in commercial broilers ΕΙΡΗΝΗ Κ. ΤΑΡΣΑΝΗ ΕΠΙΒΛΕΠΩΝ ΚΑΘΗΓΗΤΗΣ: ΑΝΤΩΝΙΟΣ ΚΟΜΙΝΑΚΗΣ Τριμελής Επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Επταμελής εξεταστική επιτροπή: Aντώνιος Κομινάκης (Αν. Καθ. ΓΠΑ) Ανδρέας Κράνης (Eρευν. B, Παν. Εδιμβούργου) Αριάδνη Χάγερ (Επ. Καθ. ΓΠΑ) Πηνελόπη Μπεμπέλη (Καθ. ΓΠΑ) Δημήτριος Βλαχάκης (Επ. Καθ. ΓΠΑ) Ευάγγελος Ζωίδης (Επ.Καθ. ΓΠΑ) Γεώργιος Θεοδώρου (Επ.Καθ. ΓΠΑ) 2 Εντοπισμός γονιδιωματικών περιοχών και δικτύων γονιδίων που επηρεάζουν παραγωγικές και αναπαραγωγικές ιδιότητες σε πληθυσμούς κρεοπαραγωγικών ορνιθίων Περίληψη Σκοπός της παρούσας διδακτορικής διατριβής ήταν ο εντοπισμός γενετικών δεικτών και υποψηφίων γονιδίων που εμπλέκονται στο γενετικό έλεγχο δύο τυπικών πολυγονιδιακών ιδιοτήτων σε κρεοπαραγωγικά ορνίθια. Μία ιδιότητα σχετίζεται με την ανάπτυξη (σωματικό βάρος στις 35 ημέρες, ΣΒ) και η άλλη με την αναπαραγωγική -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein