Expression Associated Gene Signatures of Human Epicardial Adipose Tissue
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Interactive Web Application to Explore Regeneration-Associated Gene Expression and Chromatin Accessibility
Supplementary Materials Regeneration Rosetta: An interactive web application to explore regeneration-associated gene expression and chromatin accessibility Andrea Rau, Sumona P. Dhara, Ava J. Udvadia, Paul L. Auer 1. Table S1. List of cholesterol metabolic genes from MGI database 2. Table S2. List of differentially expressed transcripts during optic nerve regeneration in zebrafish using the MGI cholesterol metabolic gene queries in the Regeneration Rosetta app 3. Table S3. List of transcription factor encoding genes from brain cell bodies following spinal cord injury in lamprey over a course of 12 weeKs 4. Table S4. List of transcription factor encoding genes from spinal cell bodies following spinal cord injury in lamprey over a course of 12 weeks Ensembl ID MGI Gene ID Symbol Name ENSMUSG00000015243 MGI:99607 Abca1 ATP-binding cassette, sub-family A (ABC1), member 1 ENSMUSG00000026944 MGI:99606 Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 ENSMUSG00000024030 MGI:107704 Abcg1 ATP binding cassette subfamily G member 1 ENSMUSG00000026003 MGI:87866 Acadl acyl-Coenzyme A dehydrogenase, long-chain ENSMUSG00000018574 MGI:895149 Acadvl acyl-Coenzyme A dehydrogenase, very long chain ENSMUSG00000038641 MGI:2384785 Akr1d1 aldo-keto reductase family 1, member D1 ENSMUSG00000028553 MGI:1353627 Angptl3 angiopoietin-like 3 ENSMUSG00000031996 MGI:88047 Aplp2 amyloid beta (A4) precursor-like protein 2 ENSMUSG00000032083 MGI:88049 Apoa1 apolipoprotein A-I ENSMUSG00000005681 MGI:88050 Apoa2 apolipoprotein A-II ENSMUSG00000032080 MGI:88051 Apoa4 -

Open Dogan Phdthesis Final.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science ELUCIDATING BIOLOGICAL FUNCTION OF GENOMIC DNA WITH ROBUST SIGNALS OF BIOCHEMICAL ACTIVITY: INTEGRATIVE GENOME-WIDE STUDIES OF ENHANCERS A Dissertation in Biochemistry, Microbiology and Molecular Biology by Nergiz Dogan © 2014 Nergiz Dogan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 ii The dissertation of Nergiz Dogan was reviewed and approved* by the following: Ross C. Hardison T. Ming Chu Professor of Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee David S. Gilmour Professor of Molecular and Cell Biology Anton Nekrutenko Professor of Biochemistry and Molecular Biology Robert F. Paulson Professor of Veterinary and Biomedical Sciences Philip Reno Assistant Professor of Antropology Scott B. Selleck Professor and Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School iii ABSTRACT Genome-wide measurements of epigenetic features such as histone modifications, occupancy by transcription factors and coactivators provide the opportunity to understand more globally how genes are regulated. While much effort is being put into integrating the marks from various combinations of features, the contribution of each feature to accuracy of enhancer prediction is not known. We began with predictions of 4,915 candidate erythroid enhancers based on genomic occupancy by TAL1, a key hematopoietic transcription factor that is strongly associated with gene induction in erythroid cells. Seventy of these DNA segments occupied by TAL1 (TAL1 OSs) were tested by transient transfections of cultured hematopoietic cells, and 56% of these were active as enhancers. Sixty-six TAL1 OSs were evaluated in transgenic mouse embryos, and 65% of these were active enhancers in various tissues. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Identification and Interaction Analysis of Molecular Markers in Pancreatic
medRxiv preprint doi: https://doi.org/10.1101/2020.12.20.20248601; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification and Interaction Analysis of Molecular Markers in Pancreatic Ductal Adenocarcinoma by Integrated Bioinformatics Analysis and Molecular Docking Experiments Basavaraj Vastrad1 , Chanabasayya Vastrad *2, Anandkumar Tengli3 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics,Chanabasava Nilaya, Bharthinagar, Dharwad, Karanataka 580001, India. 3. Department of Pharmaceutical Chemistry, JSS College of Pharmacy, Mysuru and JSS Academy of Higher Education & Research, Mysuru- 570015, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.20.20248601; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract The current investigation aimed to mine therapeutic molecular targets that play an key part in the advancement of pancreatic ductal adenocarcinoma (PDAC). The expression profiling by high throughput sequencing dataset profile GSE133684 dataset was downloaded from the Gene Expression Omnibus (GEO) database. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

An Amino-Terminal Domain of Mxil Mediates Anti-Myc Oncogenic Activity and Interacts with a Homolog of the Yeast Transcriptional Repressor SIN3
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Cell, Vol, 80, 777-786, March 10, 1995, Copyright © 1995 by Cell Press An Amino-Terminal Domain of Mxil Mediates Anti-Myc Oncogenic Activity and Interacts with a Homolog of the Yeast Transcriptional Repressor SIN3 Nicole Schreiber-Agus,*t Lynda Chin,*tt Ken Chen,t et al., 1990), and a carboxy-terminal a-helical domain re- Richard Torres, t Govinda Rao,§ Peter Guida,t quired for dimerization with another basic region-helix- Arthur h Skoultchi,§ and Ronald A. DePinhot Ioop-helix-leucine zipper (bHLH-LZ) protein, Max (Black- "rDepartments of Microbiology and Immunology wood and Eisenman, 1991; Prendergast et al., 1991). and of Medicine Many of the biochemical and biological activities of Myc §Department of Cell Biology appear to be highly dependent upon its association with ~Division of Dermatology Max (Blackwood and Eisenman, 1991 ; Prendergast et al., Albert Einstein College of Medicine 1991; Kretzner et al., 1992; Amati et al., 1993a, 1993b). Bronx, New York 10461 In addition to its key role as an obligate partner in transacti- vation-competent Myc-Max complexes, Max may also re- press Myc-responsive genes through the formation of Summary transactivation-inert complexes that are capable of bind- ing the Myc-Max recognition sequence (Blackwood et al., Documented interactions among members of the Myc 1992; Kato et al., 1992; Kretzner et al., 1992; Makela et superfamily support a yin-yang model for the regula- al., 1992; Mukherjee et al., 1992; Prendergast et al., 1992; tion of Myc-responsive genes in which t ransactivation- Ayer et al., 1993; Zervos et al., 1993). -

1 a Gene Implicated in Activation of Retinoic Acid Receptor Targets Is A
Genetics: Early Online, published on July 24, 2017 as 10.1534/genetics.117.1125 A gene implicated in activation of retinoic acid receptor targets is a novel renal agenesis gene in humans. Patrick D. Brophy, *1 Maria Rasmussen,†1 Mrutyunjaya Parida, ‡1 Greg Bonde, § Benjamin W. Darbro, * Xiaojing Hong, ‡ Jason C. Clarke, * Kevin A. Peterson, ** James Denegre, ** Michael Schneider, †† Caroline R. Sussman, ‡‡ Lone Sunde, † Dorte L. Lildballe, † Jens Michael Hertz, §§ Robert A. Cornell, §, Stephen A. Murray, ** J. Robert Manak*‡ 1Co-first authors *Department of Pediatrics, University of Iowa, Iowa City, IA, 52242. †Department of Clinical Genetics, Aarhus University Hospital, Skejby, Denmark ‡Department of Biology, University of Iowa, Iowa City, IA, 52242. §Department of Anatomy and Cell Biology, University of Iowa, IA, 52242. **The Jackson Laboratory, Bar Harbor, ME, 04609. ††Medical Genetics, Carle Foundation Hospital and Physician Group, Urbana, IL, 61801. ‡‡Department of Nephrology and Hypertension, Mayo Clinic, Rochester, MN, 55905. §§Department of Clinical Genetics, Odense University Hospital, Denmark IRB # 200711705 Whole exome sequencing data available in the Sequence Read Archive (SRA) database (accession number SRP112780) 1 Copyright 2017. Running Title: GREB1L, a novel renal agenesis gene Key Words: GREB1L, retinoic acid, renal agenesis, CAKUT, whole exome sequencing Corresponding author: J. Robert Manak, PhD 459 Biology Building, Dept. of Biology University of Iowa 129 E. Jefferson St. Iowa City, IA 52242 Ph. 319-335-0180 [email protected] 2 ABSTRACT Renal agenesis (RA) is one of the more extreme examples of congenital anomalies of the kidney and urinary tract (CAKUT). Bilateral renal agenesis is almost invariably fatal at birth, and unilateral renal agenesis can lead to future health issues including end stage renal disease. -
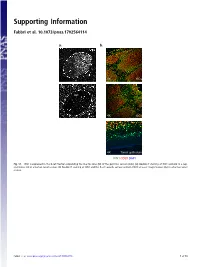
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Snps) Distant from Xenobiotic Response Elements Can Modulate Aryl Hydrocarbon Receptor Function: SNP-Dependent CYP1A1 Induction S
Supplemental material to this article can be found at: http://dmd.aspetjournals.org/content/suppl/2018/07/06/dmd.118.082164.DC1 1521-009X/46/9/1372–1381$35.00 https://doi.org/10.1124/dmd.118.082164 DRUG METABOLISM AND DISPOSITION Drug Metab Dispos 46:1372–1381, September 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Single Nucleotide Polymorphisms (SNPs) Distant from Xenobiotic Response Elements Can Modulate Aryl Hydrocarbon Receptor Function: SNP-Dependent CYP1A1 Induction s Duan Liu, Sisi Qin, Balmiki Ray,1 Krishna R. Kalari, Liewei Wang, and Richard M. Weinshilboum Division of Clinical Pharmacology, Department of Molecular Pharmacology and Experimental Therapeutics (D.L., S.Q., B.R., L.W., R.M.W.) and Division of Biomedical Statistics and Informatics, Department of Health Sciences Research (K.R.K.), Mayo Clinic, Rochester, Minnesota Received April 22, 2018; accepted June 28, 2018 ABSTRACT Downloaded from CYP1A1 expression can be upregulated by the ligand-activated aryl fashion. LCLs with the AA genotype displayed significantly higher hydrocarbon receptor (AHR). Based on prior observations with AHR-XRE binding and CYP1A1 mRNA expression after 3MC estrogen receptors and estrogen response elements, we tested treatment than did those with the GG genotype. Electrophoretic the hypothesis that single-nucleotide polymorphisms (SNPs) map- mobility shift assay (EMSA) showed that oligonucleotides with the ping hundreds of base pairs (bp) from xenobiotic response elements AA genotype displayed higher LCL nuclear extract binding after (XREs) might influence AHR binding and subsequent gene expres- 3MC treatment than did those with the GG genotype, and mass dmd.aspetjournals.org sion. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1.