Pseudomonas Syringae Pv
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Estudio Molecular De Poblaciones De Pseudomonas Ambientales
Universitat de les Illes Balears ESTUDIO MOLECULAR DE POBLACIONES DE PSEUDOMONAS AMBIENTALES T E S I S D O C T O R A L DAVID SÁNCHEZ BERMÚDEZ DIRECTORA: ELENA GARCÍA-VALDÉS PUKKITS Departamento de Biología Universitat de les Illes Balears Palma de Mallorca, Septiembre 2013 Universitat de les Illes Balears ESTUDIO MOLECULAR DE POBLACIONES DE PSEUDOMONAS AMBIENTALES Tesis Doctoral presentada por David Sánchez Bermúdez para optar al título de Doctor en el programa Microbiología Ambiental y Biotecnología, de la Universitat de les Illes Balears, bajo la dirección de la Dra. Elena García-Valdés Pukkits. Vo Bo Director de la Tesis El doctorando DRA. ELENA GARCÍA-VALDÉS PUKKITS DAVID SÁNCHEZ BERMÚDEZ Catedrática de Universidad Universitat de les Illes Balears PALMA DE MALLORCA, SEPTIEMBRE 2013 III IV Index Agradecimientos .................................................................................................... IX Resumen ................................................................................................................ 1 Abstract ................................................................................................................... 3 Introduction ............................................................................................................ 5 I.1. The genus Pseudomonas ............................................................................................ 7 I.1.1. Definition ................................................................................................................ 7 I.1.2. -

Microbiological Quality and Occurrence of Antibiotic Resistant Bacteria in Ready-To-Eat Salad
Microbiological quality and occurrence of antibiotic resistant bacteria in ready-to-eat salad Tamara Lupan Master thesis for a diploma in Food Technology and Nutrition, 30 ECTS credits Department of Food Technology, Engineering and Nutrition, Faculty of Engineering, Lund University Abstract An increase in demand for fresh vegetables resulted in an increased production of minimally processed, ready-to-eat salad in Sweden. That also brought new food safety challenges that are yet to be addressed. To assess whether ready-to-eat leafy green vegetables present a threat in terms of food borne outbreaks, aerobic bacteria, as well as bacteria belonging to the Enterobacteriaceae family, were recovered, using selective media, from 3 different sets (n=18) of ready-to-eat rocket salad. Bacterial investigation showed that most bacteria retrieved from ready-to-eat rocket salad belongs to the Pseudomonadaceae family, which are typical spoilage bacteria commonly associated with vegetables. No food-borne pathogens (e.g. Clostridium botulinum, Escherichia coli O157:H7, Salmonella spp., Shigella spp., Listeria monocytogenes) were isolated from any of three sets of rocket salad. Following the assumption that microbiological quality of the salad could change during the storage period, as well as a result of salad bags being opened, sealed, stored and opened again in the household, the total aerobic count as well as the total amount of Enterobacteriaceae were assessed every day before the best-before date. Following the NSW food Authority guidance on the microbiological status of ready-to-eat food, it can be confirmed, that each of three sets of salad were from the first day after packaging, unsatisfactory in regards to total aerobic count, showing values higher than 5 log CFU/g. -

Developing a Genetic Manipulation System for the Antarctic Archaeon, Halorubrum Lacusprofundi: Investigating Acetamidase Gene Function
www.nature.com/scientificreports OPEN Developing a genetic manipulation system for the Antarctic archaeon, Halorubrum lacusprofundi: Received: 27 May 2016 Accepted: 16 September 2016 investigating acetamidase gene Published: 06 October 2016 function Y. Liao1, T. J. Williams1, J. C. Walsh2,3, M. Ji1, A. Poljak4, P. M. G. Curmi2, I. G. Duggin3 & R. Cavicchioli1 No systems have been reported for genetic manipulation of cold-adapted Archaea. Halorubrum lacusprofundi is an important member of Deep Lake, Antarctica (~10% of the population), and is amendable to laboratory cultivation. Here we report the development of a shuttle-vector and targeted gene-knockout system for this species. To investigate the function of acetamidase/formamidase genes, a class of genes not experimentally studied in Archaea, the acetamidase gene, amd3, was disrupted. The wild-type grew on acetamide as a sole source of carbon and nitrogen, but the mutant did not. Acetamidase/formamidase genes were found to form three distinct clades within a broad distribution of Archaea and Bacteria. Genes were present within lineages characterized by aerobic growth in low nutrient environments (e.g. haloarchaea, Starkeya) but absent from lineages containing anaerobes or facultative anaerobes (e.g. methanogens, Epsilonproteobacteria) or parasites of animals and plants (e.g. Chlamydiae). While acetamide is not a well characterized natural substrate, the build-up of plastic pollutants in the environment provides a potential source of introduced acetamide. In view of the extent and pattern of distribution of acetamidase/formamidase sequences within Archaea and Bacteria, we speculate that acetamide from plastics may promote the selection of amd/fmd genes in an increasing number of environmental microorganisms. -

”Al. Beldie” Herbarium
Research Journal of Agricultural Science, 51 (3), 2019 CHARACTERIZATION OF CORNUS PLANT PRESENT IN ”AL. BELDIE” HERBARIUM Emilia VECHIU¹, Lucian DINCĂ1 1 “Marin Drăcea” National Institute for Research and Development in Forestry, Braşov, Romania email: [email protected] Abstract: ”Al. Beldie” Herbarium from ”Marin Drăcea” National Institute for Research and Development in Forestry contains a rich collection of plants. Approximately 40.000 vouchers belong to this herbarium and are stored in 600 drawers. Herbariuns are important because they provide information about plants and their area of propagation during long periods that help to carry out studies in taxonomy, biodiversity, ecology, anatomy, morphology etc. As such, various investigations were carried out with the help of data from this herbarium concerning different families and types of plants. The purpose of this article is to morphologically and ecologicallycharacterize certain Cornus species that can be found in this herbarium. Cornus Genus contains approximately 55-58 species cares that are widespread in the northern hemisphere, with few in Africa and southern America. The species found in the herbarium are the following: Cornus alba L., Cornus amomum Mill., Cornus alternifolia L., Cornus asperifolia Michx., Cornus baileyi J.M. Coult. & W.H. Evans, Cornus canadensis L., Cornus candidissima Marshall., Cornus florida L., Cornus mas L., Cornus macrophylla Wall., Cornus obliqua Raf., Cornus paniculata L'Hér., Cornus pumila Koehne, Cornus sanguinea L., Cornus stolonifera Michx. Cornus stricta Lam. and Cornus suecica L . Each plants contains data referring to the name of the species, the harvesting place, the harvesting year, the person who has collected them as well as their conservation degree. -
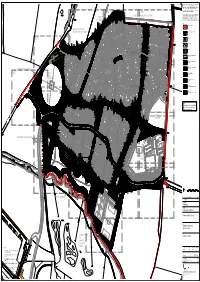
Capita Autocad Template
This drawing is the copyright of Capita. All Dimensions are to be checked not scaled. Capita accept no liability for any expense loss or damage of whatsoever nature and however arising from any variation made to this drawing or in the Specimen Feathered Feathered 337 No.Frangula alnus 1 No.Lonicera periclymenum 5 No.Sorbus aucuparia 3 No.Quercus robur execution of the work to which it relates which has not been 112 No.Corylus avellana 324 No.Ilex aquifolium 190 No.Ilex aquifolium 325 No.Frangula alnus referred to them and their approval obtained. Whip 3 No.Alnus glutinosa Feathered L-213 L-214 5 No.Alnus glutinosa L-215 Feathered 60 No.Salix caprea 3 No.Prunus avium Whip Reproduced from (insert scale of map, for example 1:10 000 3 No.Prunus avium 122 No.Corylus avellana Specimen Feathered 3 No.Salix alba scale) by permission of Ordnance Survey on behalf of The 198 No.Cornus alba 'Sibirica' EXISTING WOODLAND PLANTING 269 No.Frangula alnus 124 No.Corylus avellana Controller of Her Majesty's Stationery Office. © Crown EXISTING 223 No.Frangula alnus Copyright (insert the year of publication of the Ordnance 42 No.Salix caprea Specimen Feathered Whip 5 No.Betula pubescens 3 No.Alnus glutinosa 1 No.Lonicera periclymenum Survey data). All rights reserved. License number 100025905. HEDGE 1 No.Clematis virginiana 13 No.Corylus avellana Specimen Feathered Whip Feathered 127 No.Ilex aquifolium 5 No.Salix alba 5 No.Prunus avium 5 No.Quercus robur Specimen Feathered 157 No.Salix caprea 101 No.Cornus sanguinea 3 No.Sorbus aucuparia Feathered Whip 197 -

Sensitive and Specific Detection of Pseudomonas Avellanae Using
J. Phytopathology 149, 527±532 (2001) Ó 2001 Blackwell Wissenschafts-Verlag, Berlin ISSN 0931-1785 Istituto Sperimentale per la Frutticoltura, Ciampino Aeroporto, Roma, Italy Sensitive and Speci®c Detection of Pseudomonas avellanae using Primers based on 16S rRNA Gene Sequences M. SCORTICHINI* and U. MARCHESI Authors' address: Istituto Sperimentale per la Frutticoltura, Via di Fioranello, 52, I-00040 Ciampino Aeroporto, Rome, Italy (correspondence to M. Scortichini. E-mail: [email protected]) With 4 ®gures Received January 1, 2001; accepted May 1, 2001 Keywords: Pseudomonas avellanae, 16S rRNA gene, detection, hazelnut decline, primers Abstract up to some years. The detection of P. avellanae is A rapid polymerase chain reaction (PCR)-based proce- currently based either on traditional techniques that dure was developed for the detection of Pseudomonas include pathogenicity tests which require at least avellanae, the causal agent of hazelnut (Corylus avellana) 6±7 months for the completion or on repetitive-PCR decline in northern Greece and central Italy. The partial using the ERIC primers and requiring the isolation and sequence of the 16S rRNA gene of P. avellanae strain the production of pure cultures (Scortichini et al., PD 2390, isolated in central Italy, was compared with 2000a). The latter procedure can be completed in the sequence coding for the same gene of P. syringae pv. 4±6 days but latent infection cannot be detected. The syringae type-strain LMG 1247t1. Primers PAV 1 and possibility of utilizing a diagnostic technique enabling a PAV 22 were chosen, and after the PCR, an ampli®ca- reliable and rapid screening of the propagative material, tion product of 762 base pairs was speci®cally produced can also support the establishing of new hazelnut only by 40 strains of P. -

Cornus Sanguinea L.) Cornus Sanguinea L
Turk J Biochem 2017; 42(4): 513–518 Short Communication Zorica Popović*, Jasna Bajić-Ljubičić, Rada Matić and Srdjan Bojović First evidence and quantification of quercetin derivatives in dogberries (Cornus sanguinea L.) Cornus sanguinea L. bir tür kızılcık kuersetin türevlerinin ilk bulgusu ve miktarının DOI 10.1515/tjb-2016-0175 Conclusions: These results indicate that dogberries Received September 30, 2016; accepted April 25, 2017; previously could be a potential source of natural antioxidants, published online June 30, 2017 and encourage further investigation of this species considering that it has not yet been exploited in either Abstract nutrition or as a source of important pharmacological Aim: C. sanguinea L. is a widespread European shrubby compounds. species. It is a potential source of biologically active com- Keywords: Common dogwood; Berry extract; Flavonols; pounds, especially antioxidants, as indicated by the dog- Chemical variability. berries’ black color. The aim of the present study was to determine the content of several quercetin derivatives in the dogberries and to evaluate phytogeographical vari- Özet ability of these compounds. Materials and methods: The dogberries were collected in Amaç: C. sanguinea L, yaygın bir avrupa ağaçsı türüdür. the middle of September at two natural habitats of this Dogberries’in siyah rengiyle belirtilen biyolojik olarak species: Mt. Avala and Lake Zlatar, Serbia. The extract aktif bileşiklerin, özellikle de antioksidanların potan- obtained from fresh fruits was subjected to LC-MS/MS siyel bir kaynağıdır. Bu çalışmanın amacı, dogber- analysis to identify and quantify the content of five querce- ries çeşitli quercetin türevlerinin içeriğini belirle- tin derivatives: quercetin-3-O-glucuronide (Q-3-O-GlcA), mek ve bu bileşiklerin fitocoğrafik değişkenliklerini quercetin-3-O-galactoside (Q-3-O-Gal), quercetin-3-O-ruti- değerlendirmektir. -

Host Specificity and Virulence of the Phytopathogenic Bacteria Pseudomonas Savastanoi Eloy Caballo Ponce Caballo Eloy
Host specificity and virulence of the phytopathogenic bacteria Pseudomonas savastanoi Eloy Caballo Ponce Caballo Eloy TESIS DOCTORAL Eloy Caballo Ponce Director: Cayo Ramos Rodríguez Programa de Doctorado: Biotecnología Avanzada TESIS DOCTORAL TESIS Instituto de Hortofruticultura Subtropical y Mediterránea “La Mayora” Universidad de Málaga – CSIC 2017 Año 2017 Memoria presentada por: Eloy Caballo Ponce para optar al grado de Doctor por la Universidad de Málaga Host specificity and virulence of the phytopathogenic bacteria Pseudomonas savastanoi Director: Cayo J. Ramos Rodríguez Catedrático. Área de Genética. Departamento de Biología Celular, Genética y Fisiología. Instituto de Hortofruticultura Subtropical y Mediterránea (IHSM) Universidad de Málaga – Consejo Superior de Investigaciones Científicas Málaga, 2016 AUTOR: Eloy Caballo Ponce http://orcid.org/0000-0003-0501-3321 EDITA: Publicaciones y Divulgación Científica. Universidad de Málaga Esta obra está bajo una licencia de Creative Commons Reconocimiento-NoComercial- SinObraDerivada 4.0 Internacional: http://creativecommons.org/licenses/by-nc-nd/4.0/legalcode Cualquier parte de esta obra se puede reproducir sin autorización pero con el reconocimiento y atribución de los autores. No se puede hacer uso comercial de la obra y no se puede alterar, transformar o hacer obras derivadas. Esta Tesis Doctoral está depositada en el Repositorio Institucional de la Universidad de Málaga (RIUMA): riuma.uma.es COMITÉ EVALUADOR Presidente Dr. Jesús Murillo Martínez Departamento de Producción Agraria Universidad Pública de Navarra Secretario Dr. Francisco Manuel Cazorla López Departamento de Microbiología Universidad de Málaga Vocal Dra. Chiaraluce Moretti Departamento de Ciencias Agrarias, Alimentarias y Medioambientales Universidad de Perugia Suplentes Dr. Pablo Rodríguez Palenzuela Departamento de Biotecnología – Biología Vegetal Universidad Politécnica de Madrid Dr. -

Scientific Prospects for Cannabis-Microbiome Research To
microorganisms Review Scientific Prospects for Cannabis-Microbiome Research to Ensure Quality and Safety of Products Vladimir Vujanovic 1,*, Darren R. Korber 1 , Silva Vujanovic 2, Josko Vujanovic 3 and Suha Jabaji 4 1 Food and Bioproduct Sciences, University of Saskatchewan, Saskatoon, SK S7N 5A8, Canada; [email protected] 2 Hospital Pharmacy, CISSS des Laurentides and Université de Montréal-Montreal, QC J8H 4C7, Canada; [email protected] 3 Medical Imaging, CISSS-Laurentides, Lachute, QC J8H 4C7, Canada; [email protected] 4 Plant Science, McGill University, Ste-Anne-de-Bellevue, QC H9X 3V9, Canada; [email protected] * Correspondence: [email protected]; Tel.: +1-306-966-5048 Received: 4 December 2019; Accepted: 18 February 2020; Published: 20 February 2020 Abstract: Cannabis legalization has occurred in several countries worldwide. Along with steadily growing research in Cannabis healthcare science, there is an increasing interest for scientific-based knowledge in plant microbiology and food science, with work connecting the plant microbiome and plant health to product quality across the value chain of cannabis. This review paper provides an overview of the state of knowledge and challenges in Cannabis science, and thereby identifies critical risk management and safety issues in order to capitalize on innovations while ensuring product quality control. It highlights scientific gap areas to steer future research, with an emphasis on plant-microbiome sciences committed to using cutting-edge technologies for more efficient Cannabis production and high-quality products intended for recreational, pharmaceutical, and medicinal use. Keywords: Cannabis; microbiome; NCBI and USDA databases; omics; risk management; safety 1. Introduction The course of Cannabis science has been a top-down process, with the effects of the plant on humans and animals being tested before conducting extensive agronomic and ecological studies. -

Pseudomonas Syringae Pv. Aesculi
Pseudomonas syringae pv. aesculi Scientific Name Pseudomonas syringae pv. aesculi (ex Durgapal & Singh 1980) Young et al. 1991 Synonyms: None known Common Name(s) Bleeding canker of horse chestnut, bleeding canker disease Type of Pest Bacterial pathogen Taxonomic Position Class: Gammaproteobacteria, Order: Pseudomonadales, Family: Pseudomonadaceae Reason for Inclusion in Manual CAPS Target: Additional Pests of Concern - 2011 A B C Figure 1: A. Healthy horse chestnut tree. B. Crown dieback and yellowing associated with P. syringae pv. aesculi. C. Crown dieback and premature leaf fall seen with bleeding canker disease. Photos courtesy of the Forestry Commission. http://www.forestry.gov.uk/website/forestresearch.nsf/ByUnique/INFD- 6L4GBT Background Stem bleeding on horse chestnut trees caused by Phytophthora citricola and P. cactorum was first observed in the early 1970s in the United Kingdom and other European countries (Brasier and Strouts, 1976; Green et al., 2009). Since 2002, the 1 Last Update: February 2, 2011 number of horse chestnuts with bleeding canker, however, has dramatically increased and the disease has become severe in some parts of Europe. Initial assumptions that a Phytophthora species was the primary causal agent were refuted when the bacterium Pseudomonas syringae pv. aesculi was proven to be associated with the recent spread of the disease in the United Kingdom (Webber et al., 2007). Pest Description Pseudomonas syringae is a rod-shaped gram negative bacterium with polar flagellae (Mabbett, 2007). P. syringae exists as over 50 pathovars (strains) infecting a wide range of plants that fall within nine genomospecies (Gardan, 1999). Pathovars are genetically distinct but morphologically the same and primarily differentiated by host range. -

Comparative Analysis of the Core Proteomes Among The
Diversity 2020, 12, 289 1 of 25 Article Comparative Analysis of the Core Proteomes among the Pseudomonas Major Evolutionary Groups Reveals Species‐Specific Adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis Marios Nikolaidis 1, Dimitris Mossialos 2, Stephen G. Oliver 3 and Grigorios D. Amoutzias 1,* 1 Bioinformatics Laboratory, Department of Biochemistry and Biotechnology, University of Thessaly, 41500 Larissa, Greece; [email protected] 2 Microbial Biotechnology‐Molecular Bacteriology‐Virology Laboratory, Department of Biochemistry and Biotechnology, University of Thessaly, 41500 Larissa, Greece; [email protected] 3 Cambridge Systems Biology Centre & Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, UK; [email protected] * Correspondence: [email protected]; Tel.: +30‐2410‐565289; Fax: +30‐2410‐565290 Received: 22 June 2020; Accepted: 22 July 2020; Published: 24 July 2020 Abstract: The Pseudomonas genus includes many species living in diverse environments and hosts. It is important to understand which are the major evolutionary groups and what are the genomic/proteomic components they have in common or are unique. Towards this goal, we analyzed 494 complete Pseudomonas proteomes and identified 297 core‐orthologues. The subsequent phylogenomic analysis revealed two well‐defined species (Pseudomonas aeruginosa and Pseudomonas chlororaphis) and four wider phylogenetic groups (Pseudomonas fluorescens, Pseudomonas stutzeri, Pseudomonas syringae, Pseudomonas putida) with a sufficient number of proteomes. As expected, the genus‐level core proteome was highly enriched for proteins involved in metabolism, translation, and transcription. In addition, between 39–70% of the core proteins in each group had a significant presence in each of all the other groups. Group‐specific core proteins were also identified, with P. -

Redalyc.New Records of Lepidoptera from the Iberian Peninsula for 2015
SHILAP Revista de Lepidopterología ISSN: 0300-5267 [email protected] Sociedad Hispano-Luso-Americana de Lepidopterología España Lastuvka, A.; Lastuvka, Z. New records of Lepidoptera from the Iberian Peninsula for 2015 (Insecta: Lepidoptera) SHILAP Revista de Lepidopterología, vol. 43, núm. 172, diciembre, 2015, pp. 633-644 Sociedad Hispano-Luso-Americana de Lepidopterología Madrid, España Available in: http://www.redalyc.org/articulo.oa?id=45543699008 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative SHILAP Revta. lepid., 43 (172), diciembre 2015: 633-644 eISSN: 2340-4078 ISSN: 0300-5267 New records of Lepidoptera from the Iberian Peninsula for 2015 (Insecta: Lepidoptera) A. Lastuvka & Z. Lastuvka Abstract New records of Nepticulidae, Heliozelidae, Adelidae, Tischeriidae, Gracillariidae, Argyresthiidae, Lyonetiidae and Sesiidae for Portugal and Spain are presented. Stigmella minusculella (Herrich-Schäffer, 1855), S. tormentillella (Herrich-Schäffer, 1860), Parafomoria helianthemella (Herrich-Schäffer, 1860), Antispila metallella ([Denis & Schiffermüller], 1775), Nematopogon metaxella (Hübner, [1813]), Tischeria dodonaea Stainton, 1858, Coptotriche gaunacella (Duponchel, 1843), Caloptilia fidella (Reutti, 1853), Phyllonorycter monspessulanella (Fuchs, 1897), P. spinicolella (Zeller, 1846), Lyonetia prunifoliella