Identification of Olfactory Genes from the Greater Wax Moth by Antennal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

106Th Annual Meeting of the German Zoological Society Abstracts
September 13–16, 2013 106th Annual Meeting of the German Zoological Society Ludwig-Maximilians-Universität München Geschwister-Scholl-Platz 1, 80539 Munich, Germany Abstracts ISBN 978-3-00-043583-6 1 munich Information Content Local Organizers: Abstracts Prof. Dr. Benedikt Grothe, LMU Munich Satellite Symposium I – Neuroethology .......................................... 4 Prof. Dr. Oliver Behrend, MCN-LMU Munich Satellite Symposium II – Perspectives in Animal Physiology .... 33 Satellite Symposium III – 3D EM .......................................................... 59 Conference Office Behavioral Biology ................................................................................... 83 event lab. GmbH Dufourstraße 15 Developmental Biology ......................................................................... 135 D-04107 Leipzig Ecology ......................................................................................................... 148 Germany Evolutionary Biology ............................................................................... 174 www.eventlab.org Morphology................................................................................................ 223 Neurobiology ............................................................................................. 272 Physiology ................................................................................................... 376 ISBN 978-3-00-043583-6 Zoological Systematics ........................................................................... 416 -

Butterflies and Moths of Gwinnett County, Georgia, United States
Heliothis ononis Flax Bollworm Moth Coptotriche aenea Blackberry Leafminer Argyresthia canadensis Apyrrothrix araxes Dull Firetip Phocides pigmalion Mangrove Skipper Phocides belus Belus Skipper Phocides palemon Guava Skipper Phocides urania Urania skipper Proteides mercurius Mercurial Skipper Epargyreus zestos Zestos Skipper Epargyreus clarus Silver-spotted Skipper Epargyreus spanna Hispaniolan Silverdrop Epargyreus exadeus Broken Silverdrop Polygonus leo Hammock Skipper Polygonus savigny Manuel's Skipper Chioides albofasciatus White-striped Longtail Chioides zilpa Zilpa Longtail Chioides ixion Hispaniolan Longtail Aguna asander Gold-spotted Aguna Aguna claxon Emerald Aguna Aguna metophis Tailed Aguna Typhedanus undulatus Mottled Longtail Typhedanus ampyx Gold-tufted Skipper Polythrix octomaculata Eight-spotted Longtail Polythrix mexicanus Mexican Longtail Polythrix asine Asine Longtail Polythrix caunus (Herrich-Schäffer, 1869) Zestusa dorus Short-tailed Skipper Codatractus carlos Carlos' Mottled-Skipper Codatractus alcaeus White-crescent Longtail Codatractus yucatanus Yucatan Mottled-Skipper Codatractus arizonensis Arizona Skipper Codatractus valeriana Valeriana Skipper Urbanus proteus Long-tailed Skipper Urbanus viterboana Bluish Longtail Urbanus belli Double-striped Longtail Urbanus pronus Pronus Longtail Urbanus esmeraldus Esmeralda Longtail Urbanus evona Turquoise Longtail Urbanus dorantes Dorantes Longtail Urbanus teleus Teleus Longtail Urbanus tanna Tanna Longtail Urbanus simplicius Plain Longtail Urbanus procne Brown Longtail -

Present Or Past Herbivory: a Screening of Volatiles Released from Brassica Rapa Under Caterpillar Attacks As Attractants for the Solitary Parasitoid, Cotesia Vestalis
J Chem Ecol (2010) 36:620–628 DOI 10.1007/s10886-010-9802-6 Present or Past Herbivory: A Screening of Volatiles Released from Brassica rapa Under Caterpillar Attacks as Attractants for the Solitary Parasitoid, Cotesia vestalis Soichi Kugimiya & Takeshi Shimoda & Jun Tabata & Junji Takabayashi Received: 20 February 2010 /Revised: 7 April 2010 /Accepted: 11 May 2010 /Published online: 20 May 2010 # Springer Science+Business Media, LLC 2010 Abstract Females of the solitary endoparasitoid Cotesia decreased after removal of the host larvae, whereas vestalis respond to a blend of volatile organic compounds terpenoids and their related compounds continued to be (VOCs) released from plants infested with larvae of their released at high levels. Benzyl cyanide and dimethyl host, the diamondback moth (Plutella xylostella), which is trisulfide attracted parasitoids in a dose-dependent manner, an important pest insect of cruciferous plants. We investi- whereas the other compounds were not attractive. These gated the flight response of female parasitoids to the results suggest that nitrile and sulfide compounds tempo- cruciferous plant Brassica rapa, using two-choice tests rarily released from plants under attack by host larvae are under laboratory conditions. The parasitoids were more potentially more effective attractants for this parasitoid than attracted to plants that had been infested for at least 6 hr by other VOCs that are continuously released by host- the host larvae compared to intact plants, but they did not damaged plants. distinguish between plants infested for only 3 hr and intact plants. Although parasitoids preferred plants 1 and 2 days Key Words Herbivore-induced plant volatiles . after herbivory (formerly infested plants) over intact plants Indirect defense . -

Download Download
Agr. Nat. Resour. 54 (2020) 499–506 AGRICULTURE AND NATURAL RESOURCES Journal homepage: http://anres.kasetsart.org Research article Checklist of the Tribe Spilomelini (Lepidoptera: Crambidae: Pyraustinae) in Thailand Sunadda Chaovalita,†, Nantasak Pinkaewb,†,* a Department of Entomology, Faculty of Agriculture, Kasetsart University, Bangkok 10900, Thailand b Department of Entomology, Faculty of Agriculture at Kamphaengsaen, Kasetsart University, Kamphaengsaen Campus, Nakhon Pathom 73140, Thailand Article Info Abstract Article history: In total, 100 species in 40 genera of the tribe Spilomelini were confirmed to occur in Thailand Received 5 July 2019 based on the specimens preserved in Thailand and Japan. Of these, 47 species were new records Revised 25 July 2019 Accepted 15 August 2019 for Thailand. Conogethes tenuialata Chaovalit and Yoshiyasu, 2019 was the latest new recorded Available online 30 October 2020 species from Thailand. This information will contribute to an ongoing program to develop a pest database and subsequently to a facilitate pest management scheme in Thailand. Keywords: Crambidae, Pyraustinae, Spilomelini, Thailand, pest Introduction The tribe Spilomelini is one of the major pests in tropical and subtropical regions. Moths in this tribe have been considered as The tribe Spilomelini Guenée (1854) is one of the largest tribes and the major pests of economic crops such as rice, sugarcane, bean belongs to the subfamily Pyraustinae, family Crambidae; it consists of pods and corn (Khan et al., 1988; Hill, 2007), durian (Kuroko 55 genera and 5,929 species worldwide with approximately 86 genera and Lewvanich, 1993), citrus, peach and macadamia, (Common, and 220 species of Spilomelini being reported in North America 1990), mulberry (Sharifi et. -

Gene Families in the Desert Locust Schistocerca Gregaria
Molecular Elements Involved in Locust Olfaction: Gene Families in the Desert Locust Schistocerca gregaria Dissertation for Obtaining the Doctoral Degree of Natural Sciences (Dr. rer. nat.) Faculty of Natural Sciences University of Hohenheim From the Institute of Physiology Prof. Dr. H. Breer Submitted by Xingcong Jiang Heze (Provinz Shandong), V.R. China 2018 Dean: Prof. Dr. Uwe Beifuß 1st reviewer: Prof. Dr. Heinz Breer 2nd reviewer: Prof. Dr. Jürgen Krieger Submitted on: Oral examination on: 10.08.2018 This work was accepted by the Faculty of Natural Sciences at the University of Hohenheim as “Dissertation for Obtaining the Doctoral Degree of Natural Sciences”. Annex 2 to the University of Hohenheim doctoral degree regulations for Dr. rer. nat. Affidavit according to Sec. 7(7) of the University of Hohenheim doctoral degree regulations for Dr. rer. nat. 1. For the dissertation submitted on the topic Molecular Elements Involved in Locust Olfaction: Gene Families in the Desert Locust Schistocerca gregaria I hereby declare that I independently completed the work. 2. I only used the sources and aids documented and only made use of permissible assistance by third parties. In particular, I properly documented any contents which I used - either by directly quoting or paraphrasing - from other works. 3. I did not accept any assistance from a commercial doctoral agency or consulting firm. 4. I am aware of the meaning of this affidavit and the criminal penalties of an incorrect or incomplete affidavit. I hereby confirm the correctness of the above declaration: I hereby affirm in lieu of oath that I have, to the best of my knowledge, declared nothing but the truth and have not omitted any information. -

Occurrence of Diamondback Moths Plutella Xylostella and Their Parasitoid Wasps
bioRxiv preprint doi: https://doi.org/10.1101/357814; this version posted June 28, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Occurrence of diamondback moths Plutella xylostella and their parasitoid wasps 2 Cotesia vestalis in mizuna greenhouses and their surrounding areas 3 4 Junichiro Abe1†, Masayoshi Uefune2†, Kinuyo Yoneya3, Kaori Shiojiri4 and Junji 5 Takabayashi5 6 7 1 National Agricultural Research Center for Western Region, Ayabe, Kyoto, 623-0035, 8 Japan 9 2 Department Agrobiological Resources, Faculty of Agriculture, Meijo University, 10 Nagoya, Aichi 468-8502, Japan 11 3 Entomological Laboratory, Faculty of Agriculture, Kindai University, 3327-204, 12 Nakamachi, Nara 631-8505, Japan 13 4 Department of Agriculture, Ryukoku University, 1-5 Ooe, Otsu, Shiga 520-2194, 14 Japan 15 5 Center for Ecological Research, Kyoto University, Otsu, Shiga, 520-2113, Japan 16 17 † Both are equally contributed to this paper 18 19 Correspondence: Junichiro Abe, National Agricultural Research Center for Western 20 Region, Ayabe, Kyoto, 623-0035, Japan; Tel: +81-84-923-4100 Fax: +81-84-924-7893 21 E-mail: [email protected] 22 23 1 bioRxiv preprint doi: https://doi.org/10.1101/357814; this version posted June 28, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 24 Author Contribution 25 JA, MU and JT conceived research. A, MU, YK and KS conducted 26 experiments. JA and MU analysed field data and conducted statistical analyses. -

Identification of Genes Putatively Involved in Chitin Metabolism And
Int. J. Mol. Sci. 2015, 16, 21873-21896; doi:10.3390/ijms160921873 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article Identification of Genes Putatively Involved in Chitin Metabolism and Insecticide Detoxification in the Rice Leaf Folder (Cnaphalocrocis medinalis) Larvae through Transcriptomic Analysis Hai-Zhong Yu 1, De-Fu Wen 1, Wan-Lin Wang 2, Lei Geng 1, Yan Zhang 3 and Jia-Ping Xu 1,* 1 School of Life Sciences, Anhui Agricultural University, Hefei 230036, China; E-Mails: [email protected] (H.-Z.Y.); [email protected] (D.-F.W.); [email protected] (L.G.) 2 Rice Research Institute, Anhui Academy of Agricultural Sciences, Hefei 230031, China; E-Mail: [email protected] 3 Institute of Sericulture, Anhui Academy of Agricultural Sciences, Hefei 230061, China; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel./Fax: +86-551-6578-6691. Academic Editor: Lee A. Bulla Received: 19 May 2015 / Accepted: 25 August 2015 / Published: 10 September 2015 Abstract: The rice leaf roller (Cnaphalocrocis medinalis) is one of the most destructive agricultural pests. Due to its migratory behavior, it is difficult to control worldwide. To date, little is known about major genes of C. medinalis involved in chitin metabolism and insecticide detoxification. In order to obtain a comprehensive genome dataset of C. medinalis, we conducted de novo transcriptome sequencing which focused on the major feeding stage of fourth-instar larvae, and our work revealed useful information on chitin metabolism and insecticide detoxification and target genes of C. -

EFFECT of STORAGE DURATION on the STORED PUPAE of PARASITOID Bracon Hebetor (Say) and ITS IMPACT on PARASITOID QUALITY M
ISSN 0258-7122 (Print), 2408-8293 (Online) Bangladesh J. Agril. Res. 41(2): 297-310, June 2016 EFFECT OF STORAGE DURATION ON THE STORED PUPAE OF PARASITOID Bracon hebetor (Say) AND ITS IMPACT ON PARASITOID QUALITY M. S. ALAM1, M. Z. ALAM2, S. N. ALAM3 M. R. U. MIAH4 AND M. I. H. MIAN5 Abstract The ecto-endo larval parasitoid, Bracon hebetor (Say) is an important bio- control agent. Effective storage methods for B. hebetor are essential for raising its success as a commercial bio-control agent against lepidopteran pests. The study was undertaken to determine the effect of storage duration on the pupae of Bracon hebetor in terms of pupal survival, adult emergence, percent parasitism, female and male longevity, female fecundity and sex ratio. Three to four days old pupae were stored for 0, 1, 2, 3, 4, 5, 6, 7 and 8 weeks at 4 ± 1oC. The ranges of time for adult emergence from stored pupae, production of total adult, survivability of pupae, parasitism of host larvae by the parasitoid, longevity of adult female and male and fecundity were 63.0 -7.5 days, 6.8-43.8/50 host larvae, 13.0-99.5%, 0.0 -97.5%, 0.00-20.75 days, 0.00-17.25 days and 0.00- 73.00/50 female, respectively. The time of adult emergence and mortality of pupae increased but total number of adult emergence, survivability of pupae, longevity of adult female and male decreased gradually with the progress of storage period of B. hebetor pupae. The prevalence of male was always higher than that of female. -
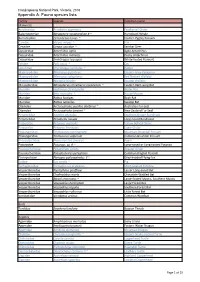
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus -
![Ichneumonid Wasps (Hymenoptera, Ichneumonidae) in the to Scale Caterpillar (Lepidoptera) [1]](https://docslib.b-cdn.net/cover/0863/ichneumonid-wasps-hymenoptera-ichneumonidae-in-the-to-scale-caterpillar-lepidoptera-1-720863.webp)
Ichneumonid Wasps (Hymenoptera, Ichneumonidae) in the to Scale Caterpillar (Lepidoptera) [1]
Central JSM Anatomy & Physiology Bringing Excellence in Open Access Research Article *Corresponding author Bui Tuan Viet, Institute of Ecology an Biological Resources, Vietnam Acedemy of Science and Ichneumonid Wasps Technology, 18 Hoang Quoc Viet, Cau Giay, Hanoi, Vietnam, Email: (Hymenoptera, Ichneumonidae) Submitted: 11 November 2016 Accepted: 21 February 2017 Published: 23 February 2017 Parasitizee a Pupae of the Rice Copyright © 2017 Viet Insect Pests (Lepidoptera) in OPEN ACCESS Keywords the Hanoi Area • Hymenoptera • Ichneumonidae Bui Tuan Viet* • Lepidoptera Vietnam Academy of Science and Technology, Vietnam Abstract During the years 1980-1989,The surveys of pupa of the rice insect pests (Lepidoptera) in the rice field crops from the Hanoi area identified showed that 12 species of the rice insect pests, which were separated into three different groups: I- Group (Stem bore) including Scirpophaga incertulas, Chilo suppressalis, Sesamia inferens; II-Group (Leaf-folder) including Parnara guttata, Parnara mathias, Cnaphalocrocis medinalis, Brachmia sp, Naranga aenescens; III-Group (Bite ears) including Mythimna separata, Mythimna loryei, Mythimna venalba, Spodoptera litura . From these organisms, which 15 of parasitoid species were found, those species belonging to 5 families in of the order Hymenoptera (Ichneumonidae, Chalcididae, Eulophidae, Elasmidae, Pteromalidae). Nine of these, in which there were 9 of were ichneumonid wasp species: Xanthopimpla flavolineata, Goryphus basilaris, Xanthopimpla punctata, Itoplectis naranyae, Coccygomimus nipponicus, Coccygomimus aethiops, Phaeogenes sp., Atanyjoppa akonis, Triptognatus sp. We discuss the general biology, habitat preferences, and host association of the knowledge of three of these parasitoids, (Xanthopimpla flavolineata, Phaeogenes sp., and Goryphus basilaris). Including general biology, habitat preferences and host association were indicated and discussed. -

Wax Worms (Galleria Mellonella) As Potential Bioremediators for Plastic Pollution Student Researcher: Alexandria Elliott Faculty Mentor: Danielle Garneau, Ph.D
Wax Worms (Galleria mellonella) as Potential Bioremediators for Plastic Pollution Student Researcher: Alexandria Elliott Faculty Mentor: Danielle Garneau, Ph.D. Center for Earth and Environmental Science SUNY Plattsburgh, Plattsburgh, NY 12901 Plastic Pollution Life History Stages Results Discussion • 30 million tons of plastic Combination of holes in plastic and • Egg stage: average length (0.478mm) • waste is generated annually and width (0.394mm) and persists 3-10 nylon in frass suggests worms are in the USA (Coalition 2018). days (Kwadha et al. 2017)(Fig. 3). digesting plastic (Figs. 6,7). • Larval stage: max length (30mm), white Of the plastic pilot trials which • 50% landfill, < 10% cream in color, possess 3 apical teeth, exhibited signs of feeding, two were HDPE (Fig. 6). recycled (PlasticsEurope, and persists 22-69 days. A Plastics The Facts 2013) • Pre-pupal/Pupal stage: length (12- • Bombelli et al. (2017) and Yang et al. 20mm) and persists 3-12 and 8-10 days, (2014) found wax worms were capable respectively. All extremities are glued to of PE consumption. • 10% of world’s plastic waste body with molting substance. Common bond (CH -CH ) in PE is 2 2 ends up in ocean 70% • Moth stage: sexual dimorphism is same as that in beeswax (Bombelli et sinks 30% floats in Fig. 1. Plastic Use distinct. Moths max length (20mm) and Fig. 5. Change in worm weight as a function of plastic pilot trial. al. 2017). Fig. 9. PE degradation as currents (Gyres, Fig. 2). (Plastics Europe). persists on average 6-14 days (males) B • Greater negative change in worm weight (g)/day for all FT-IR shows degradation of PE (i.e., evidenced by FT-IR PE and 23 days (females). -

Viruses 2015, 7, 306-319; Doi:10.3390/V7010306 OPEN ACCESS
Viruses 2015, 7, 306-319; doi:10.3390/v7010306 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review History and Current Status of Development and Use of Viral Insecticides in China Xiulian Sun Key Laboratory of Agricultural and Environmental Microbiology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China; E-Mail: [email protected]; Tel.: +86-27-8719-8641; Fax: +86-27-8719-8072 Academic Editors: John Burand and Madoka Nakai Received: 1 December 2014 / Accepted: 14 January 2015 / Published: 20 January 2015 Abstract: The use of insect viruses as biological control agents started in the early 1960s in China. To date, more than 32 viruses have been used to control insect pests in agriculture, forestry, pastures, and domestic gardens in China. In 2014, 57 products from 11 viruses were authorized as commercial viral insecticides by the Ministry of Agriculture of China. Approximately 1600 tons of viral insecticidal formulations have been produced annually in recent years, accounting for about 0.2% of the total insecticide output of China. The development and use of Helicoverpa armigera nucleopolyhedrovirus, Mamestra brassicae nucleopolyhedrovirus, Spodoptera litura nucleopolyhedrovirus, and Periplaneta fuliginosa densovirus are discussed as case studies. Additionally, some baculoviruses have been genetically modified to improve their killing rate, infectivity, and ultraviolet resistance. In this context, the biosafety assessment of a genetically modified Helicoverpa armigera nucleopolyhedrovirus is discussed. Keywords: viral insecticides; commercialization; genetic modification 1. Introduction Research on insect viruses in China started with the Bombyx mori nucleopolyhedrovirus in the mid-1950s [1] and, to date, more than 200 insect virus isolates have been recorded in China.