The Application of Gas Chromatography and Mass
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Curated Gene List for Reporting Results of Newborn Genomic Sequencing
© American College of Medical Genetics and Genomics ORIGINAL RESEARCH ARTICLE A curated gene list for reporting results of newborn genomic sequencing Ozge Ceyhan-Birsoy, PhD1,2,3, Kalotina Machini, PhD1,2,3, Matthew S. Lebo, PhD1,2,3, Tim W. Yu, MD3,4,5, Pankaj B. Agrawal, MD, MMSC3,4,6, Richard B. Parad, MD, MPH3,7, Ingrid A. Holm, MD, MPH3,4, Amy McGuire, PhD8, Robert C. Green, MD, MPH3,9,10, Alan H. Beggs, PhD3,4, Heidi L. Rehm, PhD1,2,3,10; for the BabySeq Project Purpose: Genomic sequencing (GS) for newborns may enable detec- of newborn GS (nGS), and used our curated list for the first 15 new- tion of conditions for which early knowledge can improve health out- borns sequenced in this project. comes. One of the major challenges hindering its broader application Results: Here, we present our curated list for 1,514 gene–disease is the time it takes to assess the clinical relevance of detected variants associations. Overall, 954 genes met our criteria for return in nGS. and the genes they impact so that disease risk is reported appropri- This reference list eliminated manual assessment for 41% of rare vari- ately. ants identified in 15 newborns. Methods: To facilitate rapid interpretation of GS results in new- Conclusion: Our list provides a resource that can assist in guiding borns, we curated a catalog of genes with putative pediatric relevance the interpretive scope of clinical GS for newborns and potentially for their validity based on the ClinGen clinical validity classification other populations. framework criteria, age of onset, penetrance, and mode of inheri- tance through systematic evaluation of published evidence. -

JB Review Featured Article
J. Biochem. 2011;150(3):257–266 doi:10.1093/jb/mvr090 JB Review The scent of disease: volatile organic compounds of the human body related to disease and disorder Received May 27, 2011; accepted July 6, 2011; published online July 19, 2011 Mika Shirasu and Kazushige Touhara* Human body odours also have this function; we emit a wide array of volatile organic compounds (VOCs), Department of Applied Biological Chemistry, Graduate School of both odorous and non-odorous, from our bodies. The Agricultural and Life Sciences, The University of Tokyo, Tokyo113-8657, Japan VOCs emitted from different areas of the human body vary with age, diet, sex, physiological status and pos- *Kazushige Touhara, Department of Applied Biological Chemistry, sibly genetic background. Therefore, body odours can The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, Japan 113-8657. Tel: þ03 5841 5109, Fax: þ03 5841 8024, be considered as individual ‘odour-fingerprints’. email: [email protected] Pathological processes, such as infection and en- dogenous metabolic disorders, can influence our daily Hundreds of volatile organic compounds (VOCs) are odour fingerprints by producing new VOCs or by emitted from the human body, and the components of changing the ratio of VOCs that are produced nor- VOCs usually reflect the metabolic condition of an in- mally. Therefore, it is not surprising that physicians Downloaded from dividual. Therefore, contracting an infectious or meta- have used their olfactory senses to diagnose physical bolic disease often results in a change in body odour. conditions of patients. Around 400 BD, Hippocrates Recent progresses in analytical techniques allow rapid recognized the diagnostic usefulness of body odours analyses of VOCs derived from breath, blood, skin and and reported on several disease-specific odours ema- urine. -

Test Requisition Form (Dna & Genetic Tests)
TEST REQUISITION FORM (DNA & GENETIC TESTS) INCLUDE THE FIRST 3 PAGES OF THIS FORM WITH SPECIMEN. Before sending specimens, please contact us for pre-authorization procedures. Samples received without billing pre-authorization cannot be processed. REPORTING INFORMATION ADDITIONAL REPORTS Ordering Physician or Genetic Counselor Copy of report should be sent to Name: Name: Email: Email: Institution: Fax: ( ) Address: City, State, Zip: Name: Phone: ( ) Email: Fax: ( ) Fax: ( ) PATIENT INFORMATION Patient's Last Name, First Name, MI Birthdate (mm/dd/yyyy) Gender M F Indication or reason for testing (check all that apply) Diagnosis Asymptomatic family member Confirm recorded mutation: High risk population (state Ancestry or Ethnic background below) Prenatal screening Ongoing pregnancy Other: Ancestry or Ethnic Background (check all that apply) Patient's country of origin Ethnic Background Hispanic Jewish Asian Middle-East Americas Europe Africa Australia Hospital or Clinic Patient ID Specimen ID Diagnosis (ICD9 codes) SPECIMEN INFORMATION (REQUIRED) Specimen Date/Time Collected Collected by (initial) Specimen Type (If other, please contact us before shipping) / / _____:_____ AM/PM Buccal Blood Other: Specimen may be submitted as whole blood or buccal epithelial cells (2 swabs / patient). Transport at room temperature. For other specimens and more details, see page labeled "Specimen Requirements". TESTS REQUESTED (Ask us to customize the requisition form for your practice) Test No. Test Name 1. Stat 2. Stat 3. Stat Test Names are found on the list at the end of this form. The tests will be performed in the order listed. Turnaround times are usually less than 5 weeks following receipt of specimen. Sequential panels of several large genes will take longer depending on which gene positive results are found. -

Enteral Nutrition
UnitedHealthcare® Commercial Coverage Determination Guideline Enteral Nutrition Guideline Number: CDG.027.03 Effective Date: July 1, 2021 Instructions for Use Table of Contents Page Related Commercial Policies Coverage Rationale ........................................................................... 1 • Durable Medical Equipment, Orthotics, Medical Definitions ........................................................................................... 2 Supplies and Repairs/ Replacements Applicable Codes .............................................................................. 3 References .......................................................................................10 Community Plan Policy Guideline History/Revision Information .......................................10 • Oral and Enteral Nutrition Instructions for Use .........................................................................10 Coverage Rationale Indications for Coverage Certain plans may include coverage for enteral nutrition (enteral formulas and low protein modified food products). Refer to the member specific benefit plan document to determine if this coverage applies. For Plans with Language that Cover Enteral Nutrition For plans that cover enteral nutrition, if there is a difference between a member specific benefit plan document and the information below, the member specific benefit plan document should be used for making benefit determinations. Note: Check state mandate applicability before proceeding with the following. Benefits are provided -

Genetic Testing Stories
genetic alliance monograph series number 2 Genetic testinG stories nicole exe, ms heather Ferguson, ms, cgc alyson Krokosky samantha sawyer sharon F. terry, ma published by genetic alliance Genetic testinG stories nicole exe, ms heather Ferguson, ms, cgc alyson Krokosky samantha sawyer sharon F. terry, ma published by genetic alliance 1 Genetic Alliance is grateful to the individuals and families who so generously shared their stories. They offer us a model of openness that benefits all. Partial funding for this report was provided by the Genetics and Public Policy Center. The Center is supported at Johns Hopkins University by The Pew Charitable Trusts. Any opinions expressed in this report are those of the author(s) and do not necessarily reflect the views of The Pew Chari- table Trusts or the Genetics and Public Policy Center. This report may not be reproduced or distributed in whole or in part without the permission of the Genetics and Public Policy Center. Funding for this work was also provided by the Health Resources and Services Administration, Maternal Child Health Bureau, Genetics Services Branch, cooperative agreement # 1 U33MC07945-01-00, and Genetic Alliance. 4301 connecticut ave., nW, suite 404 Washington, dc 20008 t: 202.966.5557 F: 202.966.8553 www.geneticalliance.org 2 Table of Contents PREFACE 4 HYPOKALEMIC PERIODIC PARALYSIS Adult 26 CARRIER TESTING 5 HYPOSPADIAS/CHROMOSOMAL DEFECTS Child 27 22Q11.2 DELETION SYNDROME KLINEFELTER SYNDROME Adult 27 Adult/Newborn 5 MEDIUM CHAIN ACYL COA DEHYDROGENASE ADRENOLEUKODYSTROPHY -
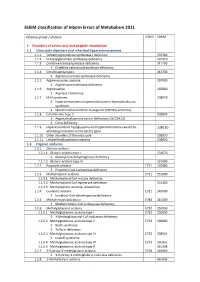
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

1 a Clinical Approach to Inherited Metabolic Diseases
1 A Clinical Approach to Inherited Metabolic Diseases Jean-Marie Saudubray, Isabelle Desguerre, Frédéric Sedel, Christiane Charpentier Introduction – 5 1.1 Classification of Inborn Errors of Metabolism – 5 1.1.1 Pathophysiology – 5 1.1.2 Clinical Presentation – 6 1.2 Acute Symptoms in the Neonatal Period and Early Infancy (<1 Year) – 6 1.2.1 Clinical Presentation – 6 1.2.2 Metabolic Derangements and Diagnostic Tests – 10 1.3 Later Onset Acute and Recurrent Attacks (Late Infancy and Beyond) – 11 1.3.1 Clinical Presentation – 11 1.3.2 Metabolic Derangements and Diagnostic Tests – 19 1.4 Chronic and Progressive General Symptoms/Signs – 24 1.4.1 Gastrointestinal Symptoms – 24 1.4.2 Muscle Symptoms – 26 1.4.3 Neurological Symptoms – 26 1.4.4 Specific Associated Neurological Abnormalities – 33 1.5 Specific Organ Symptoms – 39 1.5.1 Cardiology – 39 1.5.2 Dermatology – 39 1.5.3 Dysmorphism – 41 1.5.4 Endocrinology – 41 1.5.5 Gastroenterology – 42 1.5.6 Hematology – 42 1.5.7 Hepatology – 43 1.5.8 Immune System – 44 1.5.9 Myology – 44 1.5.10 Nephrology – 45 1.5.11 Neurology – 45 1.5.12 Ophthalmology – 45 1.5.13 Osteology – 46 1.5.14 Pneumology – 46 1.5.15 Psychiatry – 47 1.5.16 Rheumatology – 47 1.5.17 Stomatology – 47 1.5.18 Vascular Symptoms – 47 References – 47 5 1 1.1 · Classification of Inborn Errors of Metabolism 1.1 Classification of Inborn Errors Introduction of Metabolism Inborn errors of metabolism (IEM) are individually rare, but collectively numerous. -

1 Firstehg.Indd
ORIGINAL ARTICLE Egypt. J. Hum. Genet. Vol. 8, No. 2, Nov. 2007 Study of Amino Acid Disorders among a High Risk Group of Egyptian Infants and Children Rabah M. Shawky1, Heba H Elsedfy1,Amani Osman Mahmoud1, Mona Rashad1 and Eman M. Bahaa Eldin2 1Pediatric Department, Faculty of Medicine, Ain Shams University, 2Medical Genetic Centre, Ain Shams University ABSTRACT Aim of the work: The present work aimed at investigating infants (In neonatal and post neonatal period) and children suspected of having inborn errors of metabolism with unexplained mental retardation. The frequency pattern of the various amino acid disorders, in a group of selected infants and children was done to document the prevalence of various amino acid disorders among Egyptian children. Patients and Method: In this study, recent methods to investigate such disorders have been carried out by amino acid analyzer which detects levels of amino acids. Extended metabolic screen which also detects amino acid disorders, organic acid disorders and the defects of fatty acid oxidation has been carried out. These recent methods have therefore the potential of yielding information on the physiological and pathophysiological status of different metabolic pathways, as well as their interrelationship. Results: The total number of cases attending the outpatient clinic during the period of study were 1343 index cases, among them 50 index cases (3.72%) were suspected of having inborn errors of amino acid and, 20 cases (40%) of them have confirmed positive inborn errors of amino acid metabolism. Concerning the confirmed 20 cases, their ages ranged from 5 days to 11 years with a mean of 54.75±33.09 months with equally sex distribution. -

ORPHANET 3 (Phase 3)
Programme of community action on rare diseases Contract 2002/CVG4 ORPHANET : Intermediate scientific report May 2003 Summary The project was to extend the content of the already existing ORPHANET database to build up a truly European database. The first year (Dec 00-November 01) was the feasibility study year and a pilot study with four countries. The second year (Dec 01- Nov 02) was the year of the move from a French encyclopaedia to a European one, and the year of the collection of data on services in 7 countries. The third year (Dec 1,2002 – Nov 03) is the year of the data collection up to completeness in 7 of the participating countries and the year of identification of sources and satrt of the data collection in the new country: Portugal.. For the encyclopaedia, a board of 83 editors has been established progressively, specialty by specialty and authors of texts nominated. For the 3,500 diseases, there are on-line: 990 summaries in French, 833 summaries in English, 445 review articles in French or in English. The data about services are partially collected in all participating countries and already released for Italy, Belgium, Switzerland, Germany and Spain. The amount of data released is: 594 patient support groups, 945 laboratories providing diagnostic tests, 1392 research projects and 945 expert clinics. The Italian, German and Spanish versions of the website are now active. The usefulness of the database is assessed through the number of connections. In April 2003, we have had during the month visits from 101,400 different visits from 113 different countries. -

Inborn Errors of Metabolism
Inborn Errors of Metabolism Ayman W. El-Hattab, MD, FAAP, FACMG KEYWORDS Metabolic Acidosis Hypoglycemia Hyperammonemia Fatty acid oxidation Urea cycle Organic acidemia KEY POINTS Inborn errors of metabolism (IEMs) are not uncommon; their overall incidence is more than 1:1000. Neonates with IEMs usually present with nonspecific signs; therefore, maintaining a high index of suspicion is extremely important for early diagnosis of IEMs. Metabolic acidosis with hyperammonemia is suggestive of organic acidemias. Hypoglycemia without ketosis is suggestive of fatty acid oxidation defects. Hyperammonemia with respiratory alkalosis is suggestive of urea cycle defects. Liver failure can be caused by galactosemia and tyrosinemia type I. Cardiomyopathy can be caused by glycogen storage disease type II and fatty acid oxida- tion defects. Adequate caloric intake and early introduction of the appropriate enteral feeding are important in managing acute metabolic decompensation in neonates with IEMs. INTRODUCTION Inborn errors of metabolism (IEMs) are a group of disorders each of which results from deficient activity of a single enzyme in a metabolic pathway. Although IEMs are individ- ually rare, they are collectively common, with an overall incidence of more than 1:1000.1 More than 500 IEMs have been recognized, with approximately 25% of them having manifestations in the neonatal period.2,3 Neonates with IEMs are usually healthy at birth with signs typically developing in hours to days after birth. The signs are usually nonspe- cific, and may include decreased activity, poor feeding, respiratory distress, lethargy, or seizures. These signs are common to several other neonatal conditions, such as sepsis and cardiopulmonary dysfunction. Therefore, maintaining a high index of suspicion is Disclosure: None. -

Whole Exome Sequencing Gene Package Metabolic Disorders, Version 5.1, 31-1-2020
Whole Exome Sequencing Gene package Metabolic disorders, version 5.1, 31-1-2020 Technical information DNA was enriched using Agilent SureSelect DNA + SureSelect OneSeq 300kb CNV Backbone + Human All Exon V7 capture and paired-end sequenced on the Illumina platform (outsourced). The aim is to obtain 10 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate and non-unique reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA-MEM algorithm (reference: http://bio-bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x AASS Hyperlysinemia, 238700 605113 63 100 97 90 Saccharopinuria, 268700 ABAT GABA-transaminase deficiency, 613163 137150 100 100 100 96 ABCA1 HDL deficiency, familial, 1, 604091 600046 92 100 100 97 Tangier disease, 205400 -

I MERCER, HEATHER MILLIKEN, DECEMBER 2013 CELL BIOLOGY the DISTRIBUTION of SINGLE NUCLEOTIDE POLYMORPHISMS in PYODERMA GANGRENO
MERCER, HEATHER MILLIKEN, DECEMBER 2013 CELL BIOLOGY THE DISTRIBUTION OF SINGLE NUCLEOTIDE POLYMORPHISMS IN PYODERMA GANGRENOSUM: BIOMARKER DISCOVERY (124 p.) Director of Thesis: Helen Piontkivska i THE DISTRIBUTION OF SINGLE NUCLEOTIDE POLYMORPHISMS IN PYODERMA GANGRENOSUM: BIOMARKER DISCOVERY A thesis submitted To Kent State University in partial Fulfillment of the requirements for the Degree of Master of Science by Heather Milliken Mercer December 2013 ii Thesis written by Heather Milliken Mercer B.S., Kent State University, 1999 M.A., Kent State University, 2005 Approved by ___________________________________, Advisor ___________________________________, Chair, Department of Biological Sciences ___________________________________, Dean, College of Arts and Sciences iii TABLE OF CONTENTS LIST OF FIGURES……vi LIST OF TABLES……x LIST OF ABBREVIATIONS……xiii ACKNOWLEDGMENTS…………xvi CHAPTER 1: PYODERMA GANGRENOSUM, INFLAMMATION, AND APOPTOSIS…………p. 1 Hypothesis……p. 2 PG Etiology……p. 2 Multiple Pathways……p. 3 Incidence of PG……p. 4 Genetic Links to PG……p. 5 PG Clinical Variations……p. 8 Treatment of PG……p. 12 Inflammation, Apoptosis, and PG……p. 14 Errors in Apoptosis and Disease……p. 25 CHAPTER 2: METHODS…………p. 27 CHAPTER 3: RESULTS…………p. 33 CHAPTER 4: DISCUSSION…………p. 68 Significance of SNP location……p. 68 Significance of SNP state: Homozygous or Heterozygous……p. 70 PAPA Syndrome and PG SNPs in 15q24.3……p. 71 SNP_A-1874315, SNP_A-2188317, and SNP_A-2295701……p. 72 LINGO……p. 73 LOC645752……p. 74 SNP_A-1796928……p. 75 PG SNP Gene Associations, Primary Candidates, Secondary Candidates, and Master SNPs……p. 75 CASP7……p. 76 BCL2……p. 80 IL4, IL23R, IL33, IL15RA and the Interleukins……p. 82 IL15RA……p.