An Improved Taxonomic Sampling Is a Necessary but Not Sufficient Condition for Resolving Inter-Families Relationships in Caridean Decapods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Classification of Living and Fossil Genera of Decapod Crustaceans
RAFFLES BULLETIN OF ZOOLOGY 2009 Supplement No. 21: 1–109 Date of Publication: 15 Sep.2009 © National University of Singapore A CLASSIFICATION OF LIVING AND FOSSIL GENERA OF DECAPOD CRUSTACEANS Sammy De Grave1, N. Dean Pentcheff 2, Shane T. Ahyong3, Tin-Yam Chan4, Keith A. Crandall5, Peter C. Dworschak6, Darryl L. Felder7, Rodney M. Feldmann8, Charles H. J. M. Fransen9, Laura Y. D. Goulding1, Rafael Lemaitre10, Martyn E. Y. Low11, Joel W. Martin2, Peter K. L. Ng11, Carrie E. Schweitzer12, S. H. Tan11, Dale Tshudy13, Regina Wetzer2 1Oxford University Museum of Natural History, Parks Road, Oxford, OX1 3PW, United Kingdom [email protected] [email protected] 2Natural History Museum of Los Angeles County, 900 Exposition Blvd., Los Angeles, CA 90007 United States of America [email protected] [email protected] [email protected] 3Marine Biodiversity and Biosecurity, NIWA, Private Bag 14901, Kilbirnie Wellington, New Zealand [email protected] 4Institute of Marine Biology, National Taiwan Ocean University, Keelung 20224, Taiwan, Republic of China [email protected] 5Department of Biology and Monte L. Bean Life Science Museum, Brigham Young University, Provo, UT 84602 United States of America [email protected] 6Dritte Zoologische Abteilung, Naturhistorisches Museum, Wien, Austria [email protected] 7Department of Biology, University of Louisiana, Lafayette, LA 70504 United States of America [email protected] 8Department of Geology, Kent State University, Kent, OH 44242 United States of America [email protected] 9Nationaal Natuurhistorisch Museum, P. O. Box 9517, 2300 RA Leiden, The Netherlands [email protected] 10Invertebrate Zoology, Smithsonian Institution, National Museum of Natural History, 10th and Constitution Avenue, Washington, DC 20560 United States of America [email protected] 11Department of Biological Sciences, National University of Singapore, Science Drive 4, Singapore 117543 [email protected] [email protected] [email protected] 12Department of Geology, Kent State University Stark Campus, 6000 Frank Ave. -

BIOPAPUA Expedition Highlighting Deep-Sea Benthic Biodiversity of Papua New- Guinea
Biopapua Expedition – Progress report MUSÉUM NATIONAL D'HISTOIRE NATURELLE 57 rue Cuvier 75005 PARIS‐ France BIOPAPUA Expedition Highlighting deep-sea benthic Biodiversity of Papua New- Guinea Submitted by: Muséum National d'Histoire Naturelle (MNHN) Represented by (co‐PI): Dr Sarah Samadi (Researcher, IRD) Dr Philippe Bouchet (Professor, MNHN) Dr Laure Corbari (Research associate, MNHN) 1 Biopapua Expedition – Progress report Contents Foreword 3 1‐ Our understanding of deep‐sea biodiversity of PNG 4 2 ‐ Tropical Deep‐Sea Benthos program 5 3‐ Biopapua Expedition 7 4‐ Collection management 15 5‐ Preliminary results 17 6‐ Outreach and publications 23 7‐ Appendices 26 Appendix 1 27 NRI, note n°. 302/2010 on 26th march, 2010, acceptance of Biopapua reseach programme Appendix 2 28 Biopapua cruise Report, submitted by Ralph MANA (UPNG) A Report Submitted to School of Natural and Physical Sciences, University of Papua New Guinea Appendix 3 39 Chan, T.Y (2012) A new genus of deep‐sea solenocerid shrimp (Crustacea: Decapoda: Penaeoidea) from the Papua New Guinea. Journal of Crustacean Biology, 32(3), 489‐495. Appendix 4 47 Pante E, Corbari L., Thubaut J., Chan TY, Mana R., Boisselier MC, Bouchet P., Samadi S. (In Press). Exploration of the deep‐sea fauna of Papua New Guinea. Oceanography Appendix 5 60 Richer de Forges B. & Corbari L. (2012) A new species of Oxypleurodon Miers, 1886 (Crustacea Brachyura, Majoidea) from the Bismark Sea, Papua New Guinea. Zootaxa. 3320: 56–60 Appendix 6 66 Taxonomic list: Specimens in MNHN and Taiwan collections 2 Biopapua Expedition – Progress report Foreword Biopapua cruise was a MNHN/IRD deep‐sea cruise in partnership with the School of Natural and Physical Sciences, University of Papua New Guinea. -

Decapod Crustacean Grooming: Functional Morphology, Adaptive Value, and Phylogenetic Significance
Decapod crustacean grooming: Functional morphology, adaptive value, and phylogenetic significance N RAYMOND T.BAUER Center for Crustacean Research, University of Southwestern Louisiana, USA ABSTRACT Grooming behavior is well developed in many decapod crustaceans. Antennular grooming by the third maxillipedes is found throughout the Decapoda. Gill cleaning mechanisms are qaite variable: chelipede brushes, setiferous epipods, epipod-setobranch systems. However, microstructure of gill cleaning setae, which are equipped with digitate scale setules, is quite conservative. General body grooming, performed by serrate setal brushes on chelipedes and/or posterior pereiopods, is best developed in decapods at a natant grade of body morphology. Brachyuran crabs exhibit less body grooming and virtually no specialized body grooming structures. It is hypothesized that the fouling pressures for body grooming are more severe in natant than in replant decapods. Epizoic fouling, particularly microbial fouling, and sediment fouling have been shown r I m ans of amputation experiments to produce severe effects on olfactory hairs, gills, and i.icubated embryos within short lime periods. Grooming has been strongly suggested as an important factor in the coevolution of a rhizocephalan parasite and its anomuran host. The behavioral organization of grooming is poorly studied; the nature of stimuli promoting grooming is not understood. Grooming characters may contribute to an understanding of certain aspects of decapod phylogeny. The occurrence of specialized antennal grooming brushes in the Stenopodidea, Caridea, and Dendrobranchiata is probably not due to convergence; alternative hypotheses are proposed to explain the distribution of this grooming character. Gill cleaning and general body grooming characters support a thalassinidean origin of the Anomura; the hypothesis of brachyuran monophyly is supported by the conservative and unique gill-cleaning method of the group. -
W7192e19.Pdf
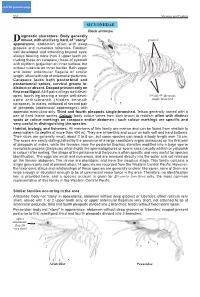
click for previous page 952 Shrimps and Prawns Sicyoniidae SICYONIIDAE Rock shrimps iagnostic characters: Body generally Drobust, with shell very hard, of “stony” grooves appearance; abdomen often with deep grooves and numerous tubercles. Rostrum well developed and extending beyond eyes, always bearing more than 3 upper teeth (in- cluding those on carapace); base of eyestalk with styliform projection on inner surface, but without tubercle on inner border. Both upper and lower antennular flagella of similar length, attached to tip of antennular peduncle. 1 Carapace lacks both postorbital and postantennal spines, cervical groove in- distinct or absent. Exopod present only on first maxilliped. All 5 pairs of legs well devel- 2 oped, fourth leg bearing a single well-devel- 3rd and 4th pleopods 4 single-branched oped arthrobranch (hidden beneath 3 carapace). In males, endopod of second pair 5 of pleopods (abdominal appendages) with appendix masculina only. Third and fourth pleopods single-branched. Telson generally armed with a pair of fixed lateral spines. Colour: body colour varies from dark brown to reddish; often with distinct spots or colour markings on carapace and/or abdomen - such colour markings are specific and very useful in distinguishing the species. Habitat, biology, and fisheries: All members of this family are marine and can be found from shallow to deep waters (to depths of more than 400 m). They are all benthic and occur on both soft and hard bottoms. Their sizes are generally small, about 2 to 8 cm, but some species can reach a body length over 15 cm. The sexes are easily distinguished by the presence of a large copulatory organ (petasma) on the first pair of pleopods of males, while the females have the posterior thoracic sternites modified into a large sperm receptacle process (thelycum) which holds the spermatophores or sperm sacs (usually whitish or yellowish in colour) after mating. -

Prawn Fauna (Crustacea: Decapoda) of India - an Annotated Checklist of the Penaeoid, Sergestoid, Stenopodid and Caridean Prawns
Available online at: www.mbai.org.in doi: 10.6024/jmbai.2012.54.1.01697-08 Prawn fauna (Crustacea: Decapoda) of India - An annotated checklist of the Penaeoid, Sergestoid, Stenopodid and Caridean prawns E. V. Radhakrishnan*1, V. D. Deshmukh2, G. Maheswarudu3, Jose Josileen 1, A. P. Dineshbabu4, K. K. Philipose5, P. T. Sarada6, S. Lakshmi Pillai1, K. N. Saleela7, Rekhadevi Chakraborty1, Gyanaranjan Dash8, C.K. Sajeev1, P. Thirumilu9, B. Sridhara4, Y Muniyappa4, A.D.Sawant2, Narayan G Vaidya5, R. Dias Johny2, J. B. Verma3, P.K.Baby1, C. Unnikrishnan7, 10 11 11 1 7 N. P. Ramachandran , A. Vairamani , A. Palanichamy , M. Radhakrishnan and B. Raju 1CMFRI HQ, Cochin, 2Mumbai RC of CMFRI, 3Visakhapatnam RC of CMFRI, 4Mangalore RC of CMFRI, 5Karwar RC of CMFRI, 6Tuticorin RC of CMFRI, 7Vizhinjam RC of CMFRI, 8Veraval RC of CMFRI, 9Madras RC of CMFRI, 10Calicut RC of CMFRI, 11Mandapam RC of CMFRI *Correspondence e-mail: [email protected] Received: 07 Sep 2011, Accepted: 15 Mar 2012, Published: 30 Apr 2012 Original Article Abstract Many penaeoid prawns are of considerable value for the fishing Introduction industry and aquaculture operations. The annual estimated average landing of prawns from the fishery in India was 3.98 The prawn fauna inhabiting the marine, estuarine and lakh tonnes (2008-10) of which 60% were contributed by freshwater ecosystems of India are diverse and fairly well penaeid prawns. An additional 1.5 lakh tonnes is produced from known. Significant contributions to systematics of marine aquaculture. During 2010-11, India exported US $ 2.8 billion worth marine products, of which shrimp contributed 3.09% in prawns of Indian region were that of Milne Edwards (1837), volume and 69.5% in value of the total export. -

Systematics, Phylogeny, and Taphonomy of Ghost Shrimps (Decapoda): a Perspective from the Fossil Record
73 (3): 401 – 437 23.12.2015 © Senckenberg Gesellschaft für Naturforschung, 2015. Systematics, phylogeny, and taphonomy of ghost shrimps (Decapoda): a perspective from the fossil record Matúš Hyžný *, 1, 2 & Adiël A. Klompmaker 3 1 Geological-Paleontological Department, Natural History Museum Vienna, Burgring 7, 1010 Vienna, Austria; Matúš Hyžný [hyzny.matus@ gmail.com] — 2 Department of Geology and Paleontology, Faculty of Natural Sciences, Comenius University, Mlynská dolina, Ilkovičova 6, SVK-842 15 Bratislava, Slovakia — 3 Florida Museum of Natural History, University of Florida, 1659 Museum Road, PO Box 117800, Gaines- ville, FL 32611, USA; Adiël A. Klompmaker [[email protected]] — * Correspond ing author Accepted 06.viii.2015. Published online at www.senckenberg.de/arthropod-systematics on 14.xii.2015. Editor in charge: Stefan Richter. Abstract Ghost shrimps of Callianassidae and Ctenochelidae are soft-bodied, usually heterochelous decapods representing major bioturbators of muddy and sandy (sub)marine substrates. Ghost shrimps have a robust fossil record spanning from the Early Cretaceous (~ 133 Ma) to the Holocene and their remains are present in most assemblages of Cenozoic decapod crustaceans. Their taxonomic interpretation is in flux, mainly because the generic assignment is hindered by their insufficient preservation and disagreement in the biological classification. Fur- thermore, numerous taxa are incorrectly classified within the catch-all taxonCallianassa . To show the historical patterns in describing fos- sil ghost shrimps and to evaluate taphonomic aspects influencing the attribution of ghost shrimp remains to higher level taxa, a database of all fossil species treated at some time as belonging to the group has been compiled: 250 / 274 species are considered valid ghost shrimp taxa herein. -

ICES Marine Science Symposia
ICES mar. Sei. Symp., 199: 209-221. 1995 Decapod crustaceans in the diets of demersal fish in the Cantabrian Sea I. Olaso and E. Rodriguez-Marin Olaso, I., and Rodriguez-Marin, E. 1995. Decapod crustaceans in the diets of demer sal fish in the Cantabrian Sea. - ICES mar. Sei. Symp., 199: 209-221. The diets of 14 species of demersal fish, representing 81% of the fish biomass of the Cantabrian Sea (ICES Division VIIIc), were determined in the spring and autumn of 1988 from the analysis of 6536 stomachs. Decapod crustaceans represented 54.3% of the diet of these fish in frequency of occurrence and 22.1% in percentage by volume. The main prey taxa were: Solenocera membranacea, Alpeus glaber, the Crangonidae and Processidae families within the Natantia, the Paguridae and Galatheidae families within the Anomura, and Goneplax rhomboides and the Portunidae family within the Brachyura. Predator-prey linkages were described and each predator was assigned to the size group that would best demonstrate any size-related feeding pattern. I. Olaso and E. Rodriguez-Marin: Instituto Espanol de Oceanografia, Laboratorio Oceanogrâfico de Santander. Apdo240, 39080 Santander, Cantabria, Spain [tel: (+34) 42 2740431275033, fax: (+34) 42 275072], Introduction Studies of size, distribution, and abundance of prey and diet selection by predators were conducted because The Bay of Biscay forms a well-defined unit because of these parameters play a fundamental role in the selec its geographical location and semi-enclosed nature. The tion of diet and have the advantage of being easily southern part of this gulf, called the Cantabrian Sea, is a quantified (Murdoch and Oaken, 1975; Vince et al., transition zone and its fish and crustacean inhabitants 1976). -

BIOLÓGICA VENEZUELICA Es Editada Por Dirección Postal De Los Mismos
7 M BIOLÓGICA II VENEZUELICA ^^.«•r-íí-yííT"1 VP >H wv* "V-i-, •^nru-wiA ">^:^;iW SWv^X/^ií. UN I VE RSIDA P CENTRAL DÉ VENEZUELA ^;."rK\'':^>:^:;':••'': ; .-¥•-^>v^:v- ^ACUITAD DE CIENCIAS INSilTÜTO DÉ Z00LOGIA TROPICAL: •RITiTRnTOrr ACTA BIOLÓGICA VENEZUELICA es editada por Dirección postal de los mismos. Deberá suministrar el Instituto de Zoología Tropical, Facultad, de Ciencias se en página aparte el título del trabajo en inglés en de la Universidad Central de Venezuela y tiene por fi caso de no estar el manuscritp elaborado en ese nalidad la publicación de trabajos originales sobre zoo idioma. logía, botánica y ecología. Las descripciones de espe cies nuevas de la flora y fauna venezolanas tendrán Resúmenes: Cada resumen no debe exceder 2 pági prioridad de publicación. Los artículos enviados no de nas tamaño carta escritas a doble espacio. Deberán berán haber sido publicados previamente ni estar sien elaborarse en castellano e ingles, aparecer en este do considerados para tal fin en otras revistas. Los ma mismo orden y en ellos deberá indicarse el objetivo nuscritos deberán elaborarse en castellano o inglés y y los principales resultados y conclusiones de la co no deberán exceder 40 páginas tamaño carta, escritas municación. a doble espacio, incluyendo bibliografía citada, tablas y figuras. Ilustraciones: Todas las ilustraciones deberán ser llamadas "figuras" y numeradas en orden consecuti ACTA BIOLÓGICA VENEZUELICA se edita en vo (Ejemplo Fig. 1. Fig 2a. Fig 3c.) el número, así co cuatro números que constituyen un volumen, sin nin mo también el nombre del autor deberán ser escritos gún compromiso de fecha fija de publicación. -

Endemic Species of Christmas Island, Indian Ocean D.J
RECORDS OF THE WESTERN AUSTRALIAN MUSEUM 34 055–114 (2019) DOI: 10.18195/issn.0312-3162.34(2).2019.055-114 Endemic species of Christmas Island, Indian Ocean D.J. James1, P.T. Green2, W.F. Humphreys3,4 and J.C.Z. Woinarski5 1 73 Pozieres Ave, Milperra, New South Wales 2214, Australia. 2 Department of Ecology, Environment and Evolution, La Trobe University, Melbourne, Victoria 3083, Australia. 3 Western Australian Museum, Locked Bag 49, Welshpool DC, Western Australia 6986, Australia. 4 School of Biological Sciences, The University of Western Australia, 35 Stirling Highway, Crawley, Western Australia 6009, Australia. 5 NESP Threatened Species Recovery Hub, Charles Darwin University, Casuarina, Northern Territory 0909, Australia, Corresponding author: [email protected] ABSTRACT – Many oceanic islands have high levels of endemism, but also high rates of extinction, such that island species constitute a markedly disproportionate share of the world’s extinctions. One important foundation for the conservation of biodiversity on islands is an inventory of endemic species. In the absence of a comprehensive inventory, conservation effort often defaults to a focus on the better-known and more conspicuous species (typically mammals and birds). Although this component of island biota often needs such conservation attention, such focus may mean that less conspicuous endemic species (especially invertebrates) are neglected and suffer high rates of loss. In this paper, we review the available literature and online resources to compile a list of endemic species that is as comprehensive as possible for the 137 km2 oceanic Christmas Island, an Australian territory in the north-eastern Indian Ocean. -

The Complete Mitogenome of €Lysmata Vittata
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.04.455109; this version posted August 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The complete mitogenome of Lysmata vittata (Crustacea: 2 Decapoda: Hippolytidae) and its phylogenetic position in 3 Decapoda 4 Longqiang Zhu1,2,3, Zhihuang Zhu1,2*, Leiyu Zhu1,2, Dingquan Wang3, Jianxin 5 Wang3*, Qi Lin1,2,3* 6 1 Fisheries Research Institute of Fujian, Xiamen, China 7 2 Key Laboratory of Cultivation and High-value Utilization of Marine Organisms in Fujian Province, Xiamen, 8 China 9 3Marine Microorganism Ecological & Application Lab, Zhejiang Ocean University, Zhejiang, China 10 * [email protected] (QL); [email protected] (JW); [email protected] (ZZ) 11 12 Abstract 13 In this study, the complete mitogenome of Lysmata vittata (Crustacea: Decapoda: 14 Hippolytidae) has been determined. The genome sequence was 22003 base pairs (bp) 15 and it included thirteen protein-coding genes (PCGs), twenty-two transfer RNA genes 16 (tRNAs), two ribosomal RNA genes (rRNAs) and three putative control regions 17 (CRs). The nucleotide composition of AT was 71.50%, with a slightly negative AT 18 skewness (-0.04). Usually the standard start codon of the PCGs was ATN, while cox1, 19 nad4L and cox3 began with TTG, TTG and GTG. The canonical termination codon 20 was TAA, while nad5 and nad4 ended with incomplete stop codon T, and cox1 ended 21 with TAG. -

Download-The-Data (Accessed on 12 July 2021))
diversity Article Integrative Taxonomy of New Zealand Stenopodidea (Crustacea: Decapoda) with New Species and Records for the Region Kareen E. Schnabel 1,* , Qi Kou 2,3 and Peng Xu 4 1 Coasts and Oceans Centre, National Institute of Water & Atmospheric Research, Private Bag 14901 Kilbirnie, Wellington 6241, New Zealand 2 Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China; [email protected] 3 College of Marine Science, University of Chinese Academy of Sciences, Beijing 100049, China 4 Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou 310012, China; [email protected] * Correspondence: [email protected]; Tel.: +64-4-386-0862 Abstract: The New Zealand fauna of the crustacean infraorder Stenopodidea, the coral and sponge shrimps, is reviewed using both classical taxonomic and molecular tools. In addition to the three species so far recorded in the region, we report Spongicola goyi for the first time, and formally describe three new species of Spongicolidae. Following the morphological review and DNA sequencing of type specimens, we propose the synonymy of Spongiocaris yaldwyni with S. neocaledonensis and review a proposed broad Indo-West Pacific distribution range of Spongicoloides novaezelandiae. New records for the latter at nearly 54◦ South on the Macquarie Ridge provide the southernmost record for stenopodidean shrimp known to date. Citation: Schnabel, K.E.; Kou, Q.; Xu, Keywords: sponge shrimp; coral cleaner shrimp; taxonomy; cytochrome oxidase 1; 16S ribosomal P. Integrative Taxonomy of New RNA; association; southwest Pacific Ocean Zealand Stenopodidea (Crustacea: Decapoda) with New Species and Records for the Region. -

Crustacea: Decapoda: Gnathophyllidae)
Memoirs of Museum Victoria 60(2): 237-242 (2003) ISSN 1447-2546 (Print) 1447-2554 (On-line) http://www.museum.vic.gov.au/memoirs Gnathophyllum taylori, a new species of caridean shrimp from south-eastern Australia (Crustacea: Decapoda: Gnathophyllidae) S h a n e T. Ah y o n g Department of Marine Invertebrates, Australian Museum, 6 College St, Sydney, NSW 2010, Australia (shanea@ austmus.gov.au) A b stra c t Ahyong, S. T. 2003. Gnathophyllum taylori, a new species of caridean shrimp from south-eastern Australia (Crustacea: Decapoda: Gnathophyllidae). Memoirs o f Museum Victoria 60(2): 237-242. Anew species of caridean shrimp, Gnathophyllum taylori sp.nov., is described from south-eastern Australia. The new species most closely resembles two Atlantic species, G. modestum Hay, 1917, and G. elegans Risso, 1816, but is readily distinguished by its banded rather than spotted body colour pattern. Gnathophyllum taylori is the third Indo-West Pacific species to be recognised. It differs from G. americanum, with which it can be sympatric, by having fewer and narrower dark bands on the body as well as by the rostral dentition, morphology of the orbit and dactyli of the walking legs. The identity of G. fasciolatum Stimpson, 1860, a junior synonym of G. americanum, is fixed by neotype selection. K eyw ords Crustacea, Decapoda, Caridea, Gnathophyllum taylori, new species Introduction Gnathophyllum taylori sp. nov. The most recent treatment of the caridean genus Figures 1-2 Gnathophyllum Latreille, 1819, recognised eight species Gnathophyllum americanum.—Healy and Yaldwyn, 1970: 46, fig. worldwide with two in the Indo-West Pacific - G.