Introduction to Nwchem
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Integrated Tools for Computational Chemical Dynamics
UNIVERSITY OF MINNESOTA Integrated Tools for Computational Chemical Dynamics • Developpp powerful simulation methods and incorporate them into a user- friendly high-throughput integrated software su ite for c hem ica l d ynami cs New Models (Methods) → Modules → Integrated Tools UNIVERSITY OF MINNESOTA Development of new methods for the calculation of potential energy surface UNIVERSITY OF MINNESOTA Development of new simulation methods UNIVERSITY OF MINNESOTA Applications and Validations UNIVERSITY OF MINNESOTA Electronic Software structu re Dynamics ANT QMMM GCMC MN-GFM POLYRATE GAMESSPLUS MN-NWCHEMFM MN-QCHEMFM HONDOPLUS MN-GSM Interfaces GaussRate JaguarRate NWChemRate UNIVERSITY OF MINNESOTA Electronic Structure Software QMMM 1.3: QMMM is a computer program for combining quantum mechanics (QM) and molecular mechanics (MM). MN-GFM 3.0: MN-GFM is a module incorporating Minnesota DFT functionals into GAUSSIAN 03. MN-GSM 626.2:MN: MN-GSM is a module incorporating the SMx solvation models and other enhancements into GAUSSIAN 03. MN-NWCHEMFM 2.0:MN-NWCHEMFM is a module incorporating Minnesota DFT functionals into NWChem 505.0. MN-QCHEMFM 1.0: MN-QCHEMFM is a module incorporating Minnesota DFT functionals into Q-CHEM. GAMESSPLUS 4.8: GAMESSPLUS is a module incorporating the SMx solvation models and other enhancements into GAMESS. HONDOPLUS 5.1: HONDOPLUS is a computer program incorporating the SMx solvation models and other photochemical diabatic states into HONDO. UNIVERSITY OF MINNESOTA DiSftDynamics Software ANT 07: ANT is a molecular dynamics program for performing classical and semiclassical trajjyectory simulations for adiabatic and nonadiabatic processes. GCMC: GCMC is a Grand Canonical Monte Carlo (GCMC) module for the simulation of adsorption isotherms in heterogeneous catalysts. -
![Automated Construction of Quantum–Classical Hybrid Models Arxiv:2102.09355V1 [Physics.Chem-Ph] 18 Feb 2021](https://docslib.b-cdn.net/cover/7378/automated-construction-of-quantum-classical-hybrid-models-arxiv-2102-09355v1-physics-chem-ph-18-feb-2021-177378.webp)
Automated Construction of Quantum–Classical Hybrid Models Arxiv:2102.09355V1 [Physics.Chem-Ph] 18 Feb 2021
Automated construction of quantum{classical hybrid models Christoph Brunken and Markus Reiher∗ ETH Z¨urich, Laboratorium f¨urPhysikalische Chemie, Vladimir-Prelog-Weg 2, 8093 Z¨urich, Switzerland February 18, 2021 Abstract We present a protocol for the fully automated construction of quantum mechanical-(QM){ classical hybrid models by extending our previously reported approach on self-parametri- zing system-focused atomistic models (SFAM) [J. Chem. Theory Comput. 2020, 16 (3), 1646{1665]. In this QM/SFAM approach, the size and composition of the QM region is evaluated in an automated manner based on first principles so that the hybrid model describes the atomic forces in the center of the QM region accurately. This entails the au- tomated construction and evaluation of differently sized QM regions with a bearable com- putational overhead that needs to be paid for automated validation procedures. Applying SFAM for the classical part of the model eliminates any dependence on pre-existing pa- rameters due to its system-focused quantum mechanically derived parametrization. Hence, QM/SFAM is capable of delivering a high fidelity and complete automation. Furthermore, since SFAM parameters are generated for the whole system, our ansatz allows for a con- venient re-definition of the QM region during a molecular exploration. For this purpose, a local re-parametrization scheme is introduced, which efficiently generates additional clas- sical parameters on the fly when new covalent bonds are formed (or broken) and moved to the classical region. arXiv:2102.09355v1 [physics.chem-ph] 18 Feb 2021 ∗Corresponding author; e-mail: [email protected] 1 1 Introduction In contrast to most protocols of computational quantum chemistry that consider isolated molecules, chemical processes can take place in a vast variety of complex environments. -

An Efficient Hardware-Software Approach to Network Fault Tolerance with Infiniband
An Efficient Hardware-Software Approach to Network Fault Tolerance with InfiniBand Abhinav Vishnu1 Manoj Krishnan1 and Dhableswar K. Panda2 Pacific Northwest National Lab1 The Ohio State University2 Outline Introduction Background and Motivation InfiniBand Network Fault Tolerance Primitives Hybrid-IBNFT Design Hardware-IBNFT and Software-IBNFT Performance Evaluation of Hybrid-IBNFT Micro-benchmarks and NWChem Conclusions and Future Work Introduction Clusters are observing a tremendous increase in popularity Excellent price to performance ratio 82% supercomputers are clusters in June 2009 TOP500 rankings Multiple commodity Interconnects have emerged during this trend InfiniBand, Myrinet, 10GigE …. InfiniBand has become popular Open standard and high performance Various topologies have emerged for interconnecting InfiniBand Fat tree is the predominant topology TACC Ranger, PNNL Chinook 3 Typical InfiniBand Fat Tree Configurations 144-Port Switch Block Diagram Multiple leaf and spine blocks 12 Spine Available in 144, 288 and 3456 port Blocks combinations 12 Leaf Blocks Multiple paths are available between nodes present on different switch blocks Oversubscribed configurations are becoming popular 144-Port Switch Block Diagram Better cost to performance ratio 12 Spine Blocks 12 Spine 12 Spine Blocks Blocks 12 Leaf 12 Leaf Blocks Blocks 4 144-Port Switch Block Diagram 144-Port Switch Block Diagram Network Faults Links/Switches/Adapters may fail with reduced MTBF (Mean time between failures) Fortunately, InfiniBand provides mechanisms to handle -

Computer-Assisted Catalyst Development Via Automated Modelling of Conformationally Complex Molecules
www.nature.com/scientificreports OPEN Computer‑assisted catalyst development via automated modelling of conformationally complex molecules: application to diphosphinoamine ligands Sibo Lin1*, Jenna C. Fromer2, Yagnaseni Ghosh1, Brian Hanna1, Mohamed Elanany3 & Wei Xu4 Simulation of conformationally complicated molecules requires multiple levels of theory to obtain accurate thermodynamics, requiring signifcant researcher time to implement. We automate this workfow using all open‑source code (XTBDFT) and apply it toward a practical challenge: diphosphinoamine (PNP) ligands used for ethylene tetramerization catalysis may isomerize (with deleterious efects) to iminobisphosphines (PPNs), and a computational method to evaluate PNP ligand candidates would save signifcant experimental efort. We use XTBDFT to calculate the thermodynamic stability of a wide range of conformationally complex PNP ligands against isomeriation to PPN (ΔGPPN), and establish a strong correlation between ΔGPPN and catalyst performance. Finally, we apply our method to screen novel PNP candidates, saving signifcant time by ruling out candidates with non‑trivial synthetic routes and poor expected catalytic performance. Quantum mechanical methods with high energy accuracy, such as density functional theory (DFT), can opti- mize molecular input structures to a nearby local minimum, but calculating accurate reaction thermodynamics requires fnding global minimum energy structures1,2. For simple molecules, expert intuition can identify a few minima to focus study on, but an alternative approach must be considered for more complex molecules or to eventually fulfl the dream of autonomous catalyst design 3,4: the potential energy surface must be frst surveyed with a computationally efcient method; then minima from this survey must be refned using slower, more accurate methods; fnally, for molecules possessing low-frequency vibrational modes, those modes need to be treated appropriately to obtain accurate thermodynamic energies 5–7. -

On the Calculation of Molecular Properties of Heavy Element Systems with Ab Initio Approaches: from Gas-Phase to Complex Systems André Severo Pereira Gomes
On the calculation of molecular properties of heavy element systems with ab initio approaches: from gas-phase to complex systems André Severo Pereira Gomes To cite this version: André Severo Pereira Gomes. On the calculation of molecular properties of heavy element systems with ab initio approaches: from gas-phase to complex systems. Theoretical and/or physical chemistry. Universite de Lille, 2016. tel-01960393 HAL Id: tel-01960393 https://hal.archives-ouvertes.fr/tel-01960393 Submitted on 19 Dec 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. M´emoirepr´esent´epour obtenir le dipl^omed' Habilitation `adiriger des recherches { Sciences Physiques de l'Universit´ede Lille (Sciences et Technologies) ANDRE´ SEVERO PEREIRA GOMES Universit´ede Lille - CNRS Laboratoire PhLAM UMR 8523 On the calculation of molecular properties of heavy element systems with ab initio approaches: from gas-phase to complex systems M´emoirepr´esent´epour obtenir le dipl^omed' Habilitation `adiriger des recherches { Sciences Physiques de l'Universit´ede Lille (Sciences et -

Starting SCF Calculations by Superposition of Atomic Densities
Starting SCF Calculations by Superposition of Atomic Densities J. H. VAN LENTHE,1 R. ZWAANS,1 H. J. J. VAN DAM,2 M. F. GUEST2 1Theoretical Chemistry Group (Associated with the Department of Organic Chemistry and Catalysis), Debye Institute, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands 2CCLRC Daresbury Laboratory, Daresbury WA4 4AD, United Kingdom Received 5 July 2005; Accepted 20 December 2005 DOI 10.1002/jcc.20393 Published online in Wiley InterScience (www.interscience.wiley.com). Abstract: We describe the procedure to start an SCF calculation of the general type from a sum of atomic electron densities, as implemented in GAMESS-UK. Although the procedure is well known for closed-shell calculations and was already suggested when the Direct SCF procedure was proposed, the general procedure is less obvious. For instance, there is no need to converge the corresponding closed-shell Hartree–Fock calculation when dealing with an open-shell species. We describe the various choices and illustrate them with test calculations, showing that the procedure is easier, and on average better, than starting from a converged minimal basis calculation and much better than using a bare nucleus Hamiltonian. © 2006 Wiley Periodicals, Inc. J Comput Chem 27: 926–932, 2006 Key words: SCF calculations; atomic densities Introduction hrstuhl fur Theoretische Chemie, University of Kahrlsruhe, Tur- bomole; http://www.chem-bio.uni-karlsruhe.de/TheoChem/turbo- Any quantum chemical calculation requires properly defined one- mole/),12 GAMESS(US) (Gordon Research Group, GAMESS, electron orbitals. These orbitals are in general determined through http://www.msg.ameslab.gov/GAMESS/GAMESS.html, 2005),13 an iterative Hartree–Fock (HF) or Density Functional (DFT) pro- Spartan (Wavefunction Inc., SPARTAN: http://www.wavefun. -
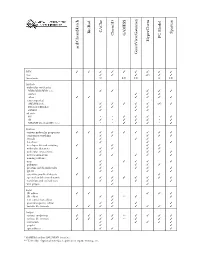
D:\Doc\Workshops\2005 Molecular Modeling\Notebook Pages\Software Comparison\Summary.Wpd
CAChe BioRad Spartan GAMESS Chem3D PC Model HyperChem acd/ChemSketch GaussView/Gaussian WIN TTTT T T T T T mac T T T (T) T T linux/unix U LU LU L LU Methods molecular mechanics MM2/MM3/MM+/etc. T T T T T Amber T T T other TT T T T T semi-empirical AM1/PM3/etc. T T T T T (T) T Extended Hückel T T T T ZINDO T T T ab initio HF * * T T T * T dft T * T T T * T MP2/MP4/G1/G2/CBS-?/etc. * * T T T * T Features various molecular properties T T T T T T T T T conformer searching T T T T T crystals T T T data base T T T developer kit and scripting T T T T molecular dynamics T T T T molecular interactions T T T T movies/animations T T T T T naming software T nmr T T T T T polymers T T T T proteins and biomolecules T T T T T QSAR T T T T scientific graphical objects T T spectral and thermodynamic T T T T T T T T transition and excited state T T T T T web plugin T T Input 2D editor T T T T T 3D editor T T ** T T text conversion editor T protein/sequence editor T T T T various file formats T T T T T T T T Output various renderings T T T T ** T T T T various file formats T T T T ** T T T animation T T T T T graphs T T spreadsheet T T T * GAMESS and/or GAUSSIAN interface ** Text only. -
![Modern Quantum Chemistry with [Open]Molcas](https://docslib.b-cdn.net/cover/4742/modern-quantum-chemistry-with-open-molcas-744742.webp)
Modern Quantum Chemistry with [Open]Molcas
Modern quantum chemistry with [Open]Molcas Cite as: J. Chem. Phys. 152, 214117 (2020); https://doi.org/10.1063/5.0004835 Submitted: 17 February 2020 . Accepted: 11 May 2020 . Published Online: 05 June 2020 Francesco Aquilante , Jochen Autschbach , Alberto Baiardi , Stefano Battaglia , Veniamin A. Borin , Liviu F. Chibotaru , Irene Conti , Luca De Vico , Mickaël Delcey , Ignacio Fdez. Galván , Nicolas Ferré , Leon Freitag , Marco Garavelli , Xuejun Gong , Stefan Knecht , Ernst D. Larsson , Roland Lindh , Marcus Lundberg , Per Åke Malmqvist , Artur Nenov , Jesper Norell , Michael Odelius , Massimo Olivucci , Thomas B. Pedersen , Laura Pedraza-González , Quan M. Phung , Kristine Pierloot , Markus Reiher , Igor Schapiro , Javier Segarra-Martí , Francesco Segatta , Luis Seijo , Saumik Sen , Dumitru-Claudiu Sergentu , Christopher J. Stein , Liviu Ungur , Morgane Vacher , Alessio Valentini , and Valera Veryazov J. Chem. Phys. 152, 214117 (2020); https://doi.org/10.1063/5.0004835 152, 214117 © 2020 Author(s). The Journal ARTICLE of Chemical Physics scitation.org/journal/jcp Modern quantum chemistry with [Open]Molcas Cite as: J. Chem. Phys. 152, 214117 (2020); doi: 10.1063/5.0004835 Submitted: 17 February 2020 • Accepted: 11 May 2020 • Published Online: 5 June 2020 Francesco Aquilante,1,a) Jochen Autschbach,2,b) Alberto Baiardi,3,c) Stefano Battaglia,4,d) Veniamin A. Borin,5,e) Liviu F. Chibotaru,6,f) Irene Conti,7,g) Luca De Vico,8,h) Mickaël Delcey,9,i) Ignacio Fdez. Galván,4,j) Nicolas Ferré,10,k) Leon Freitag,3,l) Marco Garavelli,7,m) Xuejun Gong,11,n) Stefan Knecht,3,o) Ernst D. Larsson,12,p) Roland Lindh,4,q) Marcus Lundberg,9,r) Per Åke Malmqvist,12,s) Artur Nenov,7,t) Jesper Norell,13,u) Michael Odelius,13,v) Massimo Olivucci,8,14,w) Thomas B. -

Benchmarking and Application of Density Functional Methods In
BENCHMARKING AND APPLICATION OF DENSITY FUNCTIONAL METHODS IN COMPUTATIONAL CHEMISTRY by BRIAN N. PAPAS (Under Direction the of Henry F. Schaefer III) ABSTRACT Density Functional methods were applied to systems of chemical interest. First, the effects of integration grid quadrature choice upon energy precision were documented. This was done through application of DFT theory as implemented in five standard computational chemistry programs to a subset of the G2/97 test set of molecules. Subsequently, the neutral hydrogen-loss radicals of naphthalene, anthracene, tetracene, and pentacene and their anions where characterized using five standard DFT treatments. The global and local electron affinities were computed for the twelve radicals. The results for the 1- naphthalenyl and 2-naphthalenyl radicals were compared to experiment, and it was found that B3LYP appears to be the most reliable functional for this type of system. For the larger systems the predicted site specific adiabatic electron affinities of the radicals are 1.51 eV (1-anthracenyl), 1.46 eV (2-anthracenyl), 1.68 eV (9-anthracenyl); 1.61 eV (1-tetracenyl), 1.56 eV (2-tetracenyl), 1.82 eV (12-tetracenyl); 1.93 eV (14-pentacenyl), 2.01 eV (13-pentacenyl), 1.68 eV (1-pentacenyl), and 1.63 eV (2-pentacenyl). The global minimum for each radical does not have the same hydrogen removed as the global minimum for the analogous anion. With this in mind, the global (or most preferred site) adiabatic electron affinities are 1.37 eV (naphthalenyl), 1.64 eV (anthracenyl), 1.81 eV (tetracenyl), and 1.97 eV (pentacenyl). In later work, ten (scandium through zinc) homonuclear transition metal trimers were studied using one DFT 2 functional. -

Charge-Transfer Biexciton Annihilation in a Donor-Acceptor
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2020 Supporting Information for Charge-Transfer Biexciton Annihilation in a Donor-Acceptor Co-crystal yields High-Energy Long-Lived Charge Carriers Itai Schlesinger, Natalia E. Powers-Riggs, Jenna L. Logsdon, Yue Qi, Stephen A. Miller, Roel Tempelaar, Ryan M. Young, and Michael R. Wasielewski* Department of Chemistry and Institute for Sustainability and Energy at Northwestern, Northwestern University, 2145 Sheridan Road, Evanston, Illinois 60208-3113 Contents 1. Single crystal X-ray structure data. ..................................................................................................2 2. Crystal structure determination and refinement.............................................................................3 3. Additional Steady-state absorption Spectra.....................................................................................4 4. Pump and probe spot sizes.................................................................................................................5 5. Excitation density and fraction of molecules excited calculations ..................................................6 6. Calculation of the fraction of CT excitons adjacent to one another ...............................................7 7. Calculation of reorganization energies and charge transfer rates..................................................9 8. Model Hamiltonian for calculating polarization-dependent steady-state absorption -

Quantum Chemistry (QC) on Gpus Feb
Quantum Chemistry (QC) on GPUs Feb. 2, 2017 Overview of Life & Material Accelerated Apps MD: All key codes are GPU-accelerated QC: All key codes are ported or optimizing Great multi-GPU performance Focus on using GPU-accelerated math libraries, OpenACC directives Focus on dense (up to 16) GPU nodes &/or large # of GPU nodes GPU-accelerated and available today: ACEMD*, AMBER (PMEMD)*, BAND, CHARMM, DESMOND, ESPResso, ABINIT, ACES III, ADF, BigDFT, CP2K, GAMESS, GAMESS- Folding@Home, GPUgrid.net, GROMACS, HALMD, HOOMD-Blue*, UK, GPAW, LATTE, LSDalton, LSMS, MOLCAS, MOPAC2012, LAMMPS, Lattice Microbes*, mdcore, MELD, miniMD, NAMD, NWChem, OCTOPUS*, PEtot, QUICK, Q-Chem, QMCPack, OpenMM, PolyFTS, SOP-GPU* & more Quantum Espresso/PWscf, QUICK, TeraChem* Active GPU acceleration projects: CASTEP, GAMESS, Gaussian, ONETEP, Quantum Supercharger Library*, VASP & more green* = application where >90% of the workload is on GPU 2 MD vs. QC on GPUs “Classical” Molecular Dynamics Quantum Chemistry (MO, PW, DFT, Semi-Emp) Simulates positions of atoms over time; Calculates electronic properties; chemical-biological or ground state, excited states, spectral properties, chemical-material behaviors making/breaking bonds, physical properties Forces calculated from simple empirical formulas Forces derived from electron wave function (bond rearrangement generally forbidden) (bond rearrangement OK, e.g., bond energies) Up to millions of atoms Up to a few thousand atoms Solvent included without difficulty Generally in a vacuum but if needed, solvent treated classically -

FORCE FIELDS and CRYSTAL STRUCTURE PREDICTION Contents
FORCE FIELDS AND CRYSTAL STRUCTURE PREDICTION Bouke P. van Eijck ([email protected]) Utrecht University (Retired) Department of Crystal and Structural Chemistry Padualaan 8, 3584 CH Utrecht, The Netherlands Originally written in 2003 Update blind tests 2017 Contents 1 Introduction 2 2 Lattice Energy 2 2.1 Polarcrystals .............................. 4 2.2 ConvergenceAcceleration . 5 2.3 EnergyMinimization .......................... 6 3 Temperature effects 8 3.1 LatticeVibrations............................ 8 4 Prediction of Crystal Structures 9 4.1 Stage1:generationofpossiblestructures . .... 9 4.2 Stage2:selectionoftherightstructure(s) . ..... 11 4.3 Blindtests................................ 14 4.4 Beyondempiricalforcefields. 15 4.5 Conclusions............................... 17 4.6 Update2017............................... 17 1 1 Introduction Everybody who looks at a crystal structure marvels how Nature finds a way to pack complex molecules into space-filling patterns. The question arises: can we understand such packings without doing experiments? This is a great challenge to theoretical chemistry. Most work in this direction uses the concept of a force field. This is just the po- tential energy of a collection of atoms as a function of their coordinates. In principle, this energy can be calculated by quantumchemical methods for a free molecule; even for an entire crystal computations are beginning to be feasible. But for nearly all work a parameterized functional form for the energy is necessary. An ab initio force field is derived from the abovementioned calculations on small model systems, which can hopefully be generalized to other related substances. This is a relatively new devel- opment, and most force fields are empirical: they have been developed to reproduce observed properties as well as possible. There exists a number of more or less time- honored force fields: MM3, CHARMM, AMBER, GROMOS, OPLS, DREIDING..