Lophodytes Cucull Atus): Evidence from Genetic, Mark–Recapture, and Comparative Data John M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hooded Merganser
Mergansers Order Anseriformes Family Anatinae Subfamily Mergini The Mergansers are grouped among the diving ducks but belong to their own subfamily. They are long, slender-bodied diving ducks with long, narrow saw-edged (or serrated - below, center) bills, which help them grip and hold on to fish. Often called “sawbirds,” mergansers are known for their colorful plumage and habits of flying fast & close to the water’s surface. Most species have crests on their heads, which they can hold up or down at will (top photos and below left). In flight, their head, bill, body and tail are held in a straight horizontal line. Mergansers prey on fish, fish eggs and other aquatic animals. The hooded merganser (Lod- phodytes cucullatus) - top right -lives among reeds in Pennsylvania’s swampy, woodland habi- tats using a tree cavity near a pond, lake, river or stream. They will also use a man-made nesting box placed in the proper habitat. The common merganser (Mergus merganser) - top left) needs a wilder, less inhabited site in order to nest. They too are cavity-nesters, but will also nest in a rock pile or even a hole in a stream bank (like a kingfisher). Red-breasted merganser (Mergus serrator) females build her nests in thick vegetation on the ground - though she nests in Canada and Alaska, not in Pennsylvania. But they can be seen on our open rivers during mi- gration. Merganser ducklings from various nests are often grouped to- gether and looked after by a single female.. -

P0227-P0236.Pdf
THE SEXUAL BEHAVIOR AND SYSTEMATIC POSITION OF THE HOODED MERGANSER PAUL A. JOHNSGARD T has been over 15 years since Delacour and Mayr (1945) first urged that the I mergansers (Mergzzs) and the goldeneye-Bufflehead group (Bucephala) be merged into a single tribe (Mergini) rather than being maintained in separate subfamilies (Aythyinae and Merginae) . Their reasons for this change were several, and included such points as the similarities in the downy young, female color patterns, occurrence of wild hybrids between the two genera, and tracheal structure. Indeed, except for the shape of the bill in these two groups there is no good means of distinguishing the two subfamilies. As Delacour and Mayr pointed out, bill shape and structure is highly adaptive and should not be used for the erection of major taxonomic categories. However, these two subfamilies are still upheld in the fifth edition of the AOU Check-list. Delacour and Mayr described the general similarities in the sexual behavior of lMergus and Bucephala, but no one has yet had the opportunity of critically comparing the behavior of most species in the two groups. Myres (1957,1959a, 1959b) reviewed well the behavior of the Bucephala species, but was not for- tunate enough to compare directly copulatory behavior in this genus and Mer- gus. He has, however, provided detailed descriptions of courtship and copulation in the Common Goldeneye (B. clangula) , Barrows’ Goldeneye (B. islandica) , and Bufflehead (B. albeola) . The behavior of the Common Goldeneye has also recently been described by Dane et al. (1959) and Lind (1960). I have been able to observe closely courtship display in all three species of Bucephala and in four species of Mergus, including the Hooded Merganser (M. -

2020-2021 NEBRASKA BIRD LIST See Science Olympiad General Rules, Eye Protection & Other Policies on As They Apply to Every Event
2020-2021 NEBRASKA BIRD LIST See Science Olympiad General Rules, Eye Protection & other Policies on www.soinc.org as they apply to every event Kingdom – ANIMALIA ORDER: Pelecaniformes ORDER: Gruiformes Pelicans (Pelecanidae) Rails, Gallinules, and Coots (Rallidae) Phylum – CHORDATA American White Pelican Pelecanus Clapper Rail Rallus longirostris Subphylum – VERTEBRATA erythrorhynchos Sora Bitterns, Herons, and Allies Purple Gallinule Class - AVES (Ardeidae) American Coot Family Group (Family Name) American Bittern Cranes (Gruidae) Common Name Great Blue Heron Ardea herodias Sandhill Crane Antigone canadensis [Scientific name is in italics] Snowy Egret Egretta thula Whooping Crane Grus americana Green Heron ORDER: Anseriformes Black-crowned Night-heron ORDER: Charadriiformes Ducks, Geese, and Swans (Anatidae) Ibises and Spoonbills Lapwings and Plovers (Charadriidae) Black-bellied Whistling-duck (Threskiornithidae) American Golden-Plover Snow Goose Roseate Spoonbill Platalea ajaja Piping Plover Charadrius melodus Canada Goose Branta canadensis Killdeer Charadrius vociferus Trumpeter Swan ORDER: Suliformes Oystercatchers (Haematopodidae) Wood Duck Aix sponsa Cormorants (Phalacrocoracidae) American Oystercatcher Mallard Anas platyrhynchos Double-crested Cormorant Stilts and Avocets (Recurvirostridae) Northern Shoveler Phalacrocorax auritus Black-necked Stilt Green-winged Teal Darters (Anhingida) American Avocet Recurvirostra Canvasback Anhinga Anhinga anhinga americana Hooded Merganser Frigatebirds (Fregatidae) Sandpipers, Phalaropes, -

Diseases of Seaducks in Captivity
Diseases of seaducks in captivity NIGELLA HILLGARTH and JANET KEAR For many years the Wildfowl Trust has Bufflehead Bucephaia albeola are, on the monitored the health of its collections by other hand, notoriously difficult to breed carrying out post-mortem examinations. (Williams 1971), and the Hooded Merganser Detailed records have been kept by J. V. M. cucullatus is only a little easier (Lubbock Beer (1959—1969), by N. A. Wood 1973). (1970-1973), and by M. J. Brown sub The difficulty with which these ducks are sequently. This analysis of the accumulated bred is, to some extent, reflected in the records is the first of a series that will numbers available from the Wildfowl Trust’s describe the cause of death in the various collections at post-mortem. For instance, wildfowl groups. eiders predominate in the records, and saw There are seven genera, and 18 species, of bills are commoner than scoters. However, seaducks (M ergini) including the eiders eggs have been collected on a number of oc (Johnsgard 1960). All except one species in casions from the wild, and hatched artificial habit the higher latitudes of the northern ly, so post-mortem findings from dead dow hemisphere; the rare exception, the Brazilian ny and juvenile birds do exist, even for M erganser Mergus octosetaceus, has never species that have seldom bred in captivity. been maintained for long in captivity. The whole group is generally difficult to keep and to persuade to breed. There are many M aterials reasons for this; seaducks are divers and animal eaters; most of them spend the Post-mortem data from 641 seaducks dying winter, and some breed, on salt water; those between 1959 and 1976 have been examined. -

List of Native and Naturalized Fauna of Virginia
Virginia Department of Wildlife Resources List of Native and Naturalized Fauna of Virginia August, 2020 (* denotes naturalized species; ** denotes species native to some areas of Virginia and naturalized in other areas of Virginia) Common Name Scientific Name FISHES: Freshwater Fishes: Alabama Bass * Micropterus henshalli * Alewife Alosa pseudoharengus American Brook Lamprey Lampetra appendix American Eel Anguilla rostrata American Shad Alosa sapidissima Appalachia Darter Percina gymnocephala Ashy Darter Etheostoma cinereum Atlantic Sturgeon Acipenser oxyrhynchus Banded Darter Etheostoma zonale Banded Drum Larimus fasciatus Banded Killifish Fundulus diaphanus Banded Sculpin Cottus carolinae Banded Sunfish Ennaecanthus obesus Bigeye Chub Hybopsis amblops Bigeye Jumprock Moxostoma ariommum Bigmouth Chub Nocomis platyrhynchus Black Bullhead Ameiurus melas Black Crappie Pomoxis nigromaculatus Blacktip Jumprock Moxostoma cervinum Black Redhorse Moxostoma duquesnei Black Sculpin Cottus baileyi Blackbanded Sunfish Enneacanthus chaetodon Blacknose Dace Rhinichthys atratulus Blackside Dace Chrosomus cumberlandensis Blackside Darter Percina maculata Blotched Chub Erimystax insignis Blotchside Logperch Percina burtoni Blue Catfish * Ictalurus furcatus * Blue Ridge Sculpin Cottus caeruleomentum Blueback Herring Alosa aestivalis Bluebreast Darter Etheostoma camurum Bluegill Lepomis macrochirus Bluehead Chub Nocomis leptocephalus Blueside Darter Etheostoma jessiae Bluespar Darter Etheostoma meadiae Bluespotted Sunfish Enneacanthus gloriosus Bluestone -

PHYLOGENY and EVOLUTIONARY ECOLOGY of MODERN SEADUCKS (ANATIDAE: MERGINI)L
The Condor97:233-255 0 The Ccaper OrnithologicalSociety 1995 PHYLOGENY AND EVOLUTIONARY ECOLOGY OF MODERN SEADUCKS (ANATIDAE: MERGINI)l BRADLEY C. LIVEZEY Section of Birds, CarnegieMuseum of Natural History, 4400 ForbesAvenue, Pittsburgh,PA 15213-4080 Abstract. Phylogenetic relationships of modem seaducks(Me&i) were investigated using a cladistic analysis of 137 morphological characters.The analysis produced a single tree (consistencyindex = 0.692, excludingautapomorphies) with complete resolutionof the relationshipsamong the 25 taxa recognized. Phylogeneticinferences include: (1) the eiders (Polystictaand Somateria) constitutea monophyletic group and are the sister-groupof other Mergini; (2) the remaining generic groups of Mergini, sequencedin order of increasingly close relationship, are Histrionicus,Melanitta + Camptorhynchus,Clangula, Bucephala + Mergellus.Lophodytes, and Mergus;(3) Somateria is monophyletic with S. jischerithe sister- group of its congeners;(4) the black scoters(Melanitta nigra-group)are the sister-groupof other scoters;(5) the Bufflehead (Bucephalaalbeola) is the sister-groupof the goldeneyes (B. clangufaand B. islandica);and (6) relationshipsamong mergansersare as presentedby Livezey (1989). Bootstrappingrevealed that the placement of the Smew as the sister-group of the goldeneyesis only weakly supported,and ancillary analysesrevealed that this place- ment is only one step shorter than its reduction to a trichotomy with the goldeneyesand mergansers(including Lophodytes).A phylogenetic classification of modem seaducksis -
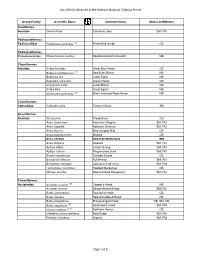
Bird List 4A.Xlsx
List of Birds Observed at the Midewin National Tallgrass Prairie Group/Family Scientific Name Common Name Status At Midewin Gaviiformes Gaviidae Gavia immer Common Loon SM, FM Podicipediformes Podicipedidae Podilymbus podiceps (T) Pied-billed Grebe CB Podicipediformes Phalacrocoracidae Phalacrocorax auritus Doublecrested Cormorant NB Ciconiiformes Ardeidae Ardea herodias Great Blue Heron CB Botaurus lentiginosus (E) American Bittern NB Bubulcus ibis Cattle Egret NB Butorides virescens Green Heron CB Ixobrychus exilis Least Bittern CB Ardea alba Great Egret NB Nycticorax nycticorax (E) Black-crowned Night-Heron NB Ciconiiformes Cathartidae Cathartes aura Turkey Vulture SB Anseriformes Anatidae Aix sponsa Wood Duck CB Anas americana American Wigeon SM, FM Anas clypeata Northern Shoveler SM, FM Anas discors Blue-winged Teal CB Anas platyrhynchos Mallard CB Anas rubripes American Black Duck SM Anas strepera Gadwall SM, FM Aythya affinis Lesser Scaup SM, FM Aythya collaris Ring-necked Duck SM, FM Branta canadensis Canada Goose CB Bucephala albeola Bufflehead SM, FM Bucephala clangula Common Goldeneye SM, FM Lophodytes cucullatus Hooded Merganser CB Mergus serrator Red-breasted Merganser SM, FM Falconiformes Accipitridae Accipiter cooperii (E) Cooper's Hawk NB Accipiter striatus Sharp-shinned Hawk SM, FM Buteo jamaicensis Red-tailed Hawk CB Buteo lineatus Red-shouldered Hawk NB Buteo platypterus Broad-winged Hawk NB, SM, FM Buteo swainsoni (E) Swainson's Hawk SM, FM Circus cyaneus (E,A) Northern Harrier CB Haliaeetus leucocephalus Bald Eagle -

ANSERIFORMES Taxon Advisory Group Regional Collection Plan 3Rd Edition • 2020 - 2025
ANSERIFORMES Taxon Advisory Group Regional Collection Plan 3rd Edition • 2020 - 2025 Edited by Photo by Pinola Conservancy Keith Lovett, Anseriformes TAG Chair Buttonwood Park Zoo Table of Contents Acknowledgements 03 TAG Operational Structure 04 TAG Steering Committee and Advisors: Table 1 05 TAG Definition 06 TAG Mission 06 TAG Vision 06 TAG Strategic Planning Overview 07 TAG Goals Sustainability 08 Conservation 09 Husbandry and Welfare 09 Educational Waterfowl Awareness and Program Support 10 TAG Taxonomy 11 TAG Taxonomy: Table 2 12 Conservation Status of Anseriformes Overview 13 Conservation Status of Anseriformes: Table 3 14 RCP History and Program Designation Program Management Designation 20 Additional Management Designation 21 Selection Criteria Selection Criteria Overview 22 Decision Tree Selection Criteria Categories 23 Proposed EAZA Waterfowl TAG European Endangered 24 Species Program (EEP) Species Anseriformes Decision Tree: Table 4 25 Anseriformes TAG Selection Criteria / Decision Tree: Table 5 26 Anseriformes TAG Selection Criteria / Decision Tree Summary 33 Space Assessment Space Assessment Overview 34 Space Survey Accountability:Responding Insititutions: Table 6 35 Space Survey Accountability: Non-responding Insititutions: Table 7 37 Space Survey Results and Target Size: Table 8 38 Regional Anseriformes Populations: Table 9 39 Summary Table 46 Non-Recommended Species Replacement Overview 47 Non-Recommended Species Replacement Chart: Table 11 48 Management Update: Table 12 51 SSP Five Year Goals and Essential Actions 53 -
Birds of Toronto
i BIRDS OF TORONTO A GUIDE TO THEIR REMARKABLE WORLD • City of Toronto Biodiversity Series • Imagine a Toronto with flourishing natural habitats and an urban environment made safe for a great diversity of wildlife species. Envision a city whose residents treasure their daily encounters with the remarkable and inspiring world of nature, and the variety of plants and animals who share this world. Take pride in a Toronto that aspires to be a world leader in the development of urban initiatives that will be critical to the preservation of our flora and fauna. Cover photo: Jean Iron A flock of Whimbrel viewed from Colonel Samuel Smith Park on 23 May 2007 frames the Toronto skyline. Since the early 20th century, Toronto ornithologists have noted the unique and impressive spring migration of Whimbrel past the city’s waterfront within a narrow 22 – 27 May time frame. In this short stretch of May, literally thousands of Whimbrel migrate past Toronto each spring between their South American wintering grounds and their breeding grounds on the tundra coast of the Hudson Bay Lowlands. In some years, as much as one quarter of the entire eastern North American population is witnessed passing along the Lake Ontario shoreline. Afforded protection by the Migratory Birds Convention Act of 1917, its population is probably still rebounding from intense market hunting pressure in the 19th century. City of Toronto © 2011 American Woodcock ISBN 978-1-895739-67-1 Barry Kent MacKay 1 “Indeed, in its need for variety and acceptance of randomness, a flourishing TABLE OF CONTENTS natural ecosystem is more like a city than like a plantation. -

2016 Canadian NAWMP Report
September 2016 nawmp.wetlandnetwork.ca HabitatMatters 2016 Canadian NAWMP Report “Offshore Wind – Surf Scoters” from the 2016 Canadian Wildlife Habitat Conservation Stamp series. Artist: Pierre Leduc, Stoneham (QC) North American Waterfowl Management Plan —— Plan nord-américain de gestion de la sauvagine —— Plan de Manejo de Aves 2016 Canadian North American Waterfowl Management Plan Report AcuáticasHabitat MattersNorteaméricaa TableContents of 1 About the NAWMP 2 National Overview 2 Accomplishments 3 Expenditures and Contributions 4 Special Feature – A Year of Milestones 6 Habitat Joint Ventures 7 Pacific Birds Habitat Joint Venture 14 Canadian Intermountain Joint Venture 18 Prairie Habitat Joint Venture 24 Eastern Habitat Joint Venture 28 Species Joint Ventures 29 Black Duck Joint Venture 32 Sea Duck Joint Venture 34 Arctic Goose Joint Venture 36 Partners b Habitat Matters 2016 Canadian North American Waterfowl Management Plan Report About the NAWMP Mallards landing on water. The North American Waterfowl Management Plan (NAWMP or Ducks Unlimited Canada ‘the Plan’) is an international partnership to restore, conserve and protect waterfowl populations and associated habitats through management decisions based on strong biological waterfowl populations. Mexico became a signatory to foundations. The ultimate goal is to achieve abundant and the Plan with its update in 1994. As a result, the NAWMP resilient waterfowl populations and sustainable landscapes. partnership extends across North America, working at national The Plan engages the community of users and supporters and regional levels on a variety of waterfowl and habitat committed to conservation and valuing waterfowl. management issues. In 1986, the Canadian and American governments signed this Since the creation of the Plan, NAWMP partners have worked international partnership agreement, laying the foundation to conserve and restore wetlands, associated uplands and for international cooperation in the recovery of declining other key habitats for waterfowl across Canada, the United States and Mexico. -
WILDLIFE National Wildlife Refuge
PEA ISLAND WILDLIFE National Wildlife Refuge • General Information • Amphibians • Reptiles • Mammals • Birds • Birds • Birds General Pea Island National Wildlife Refuge, located in Dare County, North Carolina, is one of over 400 national wildliferefuges nationwide administered by the U.S. Fish and Wildlife Service. Though each refuge was established to provide habitat for certain primary species, each is managed to provide for a diversity of wildlife and opportunities for public enjoyment of these natural resources. Wildlife observation is a popular pastime for millions of people each year. This publication is designed to help refuge visitors observe and identify the wildlife of Pea Island National Wildlife Refuge. Established in 1938 as a wintering sanctuary for waterfowl, Pea Island contains 5,915 acres of coastal barrier island and 25,700 acres of Proclamation Boundary Waters in the Pamlico Sound. Management of the refuge provides diverse habitat types including beach, dunes, salt marsh, fresh and brackish water ponds and salt flats. , In observing the 'Yildlife populations here, remember the dynamic nature of a barrier island. Over the years inlets open and close. The ocean overwashed inter mittently during frequent winter storms. Sometimes all surface water is salty. Animal populations here generally have more limiting factors than other places. On an island the mere existence of a species may depend soleiy on its ability to swim or fly. Survival largely relates to its ability to tolerate the harsh salt environment. Is it any wonder that our richest diversity of wildlife is among the avian popu lations and our least diverse, the amphibians, who would dehydrate in salt water? Ponder these environ mental factors as you observe refuge wildlife. -

Birds List for Carlsbad Caverns Np
BIRDS LIST FOR CARLSBAD CAVERNS NP Order Family Scientific Name Common Name PODICIPEDIFORMES Podicipedidae Aechmophorus occidentalis Western Grebe PODICIPEDIFORMES Podicipedidae Podiceps nigricollis Eared Grebe PODICIPEDIFORMES Podicipedidae Podilymbus podiceps Pied-billed Grebe PELECANIFORMES Pelecanidae Pelecanus erythrorhynchos American White Pelican PELECANIFORMES Phalacrocoracidae Phalacrocorax auritus Double-crested Cormorant CICONIIFORMES Ardeidae Ardea herodias Great Blue Heron CICONIIFORMES Ardeidae Botaurus lentiginosus American Bittern CICONIIFORMES Ardeidae Bubulcus ibis Cattle Egret CICONIIFORMES Ardeidae Butorides striatus Green-backed Heron CICONIIFORMES Ardeidae Casmerodius albus Great Egret CICONIIFORMES Ardeidae Egretta thula Snowy Egret CICONIIFORMES Ardeidae Egretta tricolor Tricolored Heron CICONIIFORMES Ardeidae Nycticorax nycticorax Black-crowned Night-Heron CICONIIFORMES Threskiornithidae Eudocimus albus White Ibis CICONIIFORMES Threskiornithidae Plegadis chihi White-faced Ibis ANSERIFORMES Anatidae Aix sponsa Wood Duck ANSERIFORMES Anatidae Anas acuta Northern Pintail ANSERIFORMES Anatidae Anas americana American Wigeon ANSERIFORMES Anatidae Anas clypeata Northern Shoveler ANSERIFORMES Anatidae Anas crecca Green-winged Teal ANSERIFORMES Anatidae Anas cyanoptera Cinnamon Teal ANSERIFORMES Anatidae Anas discors Blue-winged Teal ANSERIFORMES Anatidae Anas platyrhynchos Mallard ANSERIFORMES Anatidae Anas strepera Gadwall ANSERIFORMES Anatidae Aythya affinis Lesser Scaup ANSERIFORMES Anatidae Aythya americana Redhead