Encyclopedia of Plant Viruses and Viroids K
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alfalfa PMG 12 19 11
UC IPM Pest Management Guidelines: ALFALFA September 2010 Contents (Dates in parenthesis indicate when each topic was updated) Alfalfa Year-Round IPM Program Checklist (11/06) ........................................................................................................................ iv General Information Integrated Pest Management (11/06) ................................................................................................................................................... 1 Selecting the Field (11/06) ................................................................................................................................................................... 2 Transgenic Herbicide-Tolerant Alfalfa (11/06) ................................................................................................................................... 3 Biological Control (11/06) ................................................................................................................................................................... 5 Sampling with a Sweep Net (11/06) .................................................................................................................................................... 6 Crop Rotation (11/06) .......................................................................................................................................................................... 8 Aphid Monitoring (9/07) ..................................................................................................................................................................... -

Thrips-Transmitted Tomato Spotted Wilt Virus (TSWV) in California Crops
Emergence and integrated management of thrips-transmitted Tomato spotted wilt virus (TSWV) in California crops UCCE-Monterey County; 2015 Plant Disease Seminar November 4, 2015; County of Monterey Agricultural Center; Salinas, California Ozgur Batuman Department of Plant Pathology, UC Davis California Processing Tomatoes And Peppers • Today, California grows 95 percent of the USA’s processing tomatoes and approximately 30 percent of the world processing tomatoes production! • California produced 60 and 69 percent of the bell peppers and chile peppers, respectively, grown in the USA in 2014! Pepper-infecting viruses • ~70 viruses known to infect peppers worldwide • ~10 of these known to occur in California • Most are not seed-transmitted • Difficult to identify based on symptoms • Mixed infections are common • Transmitted from plant-to-plant by various insects, primarily aphids and thrips • Best managed by an IPM approach TSWV Key viruses affecting peppers in CA production areas • Alfalfa mosaic virus (AMV) Alfamovirus • Cucumber mosaic virus (CMV) Cucumovirus • Pepper mottle virus (PepMoV) Potyvirus Aphid • Potato virus Y (PVY) Potyvirus • Tobacco etch virus (TEV) Potyvirus • Pepper mild mottle virus (PMMV) Tobamovirus • Tobacco mosaic virus (TMV) Tobamovirus Seed & Mechanical • Tomato mosaic virus (ToMV) Tobamovirus • Tomato spotted wilt virus (TSWV) Tospovirus Thrips • Impatient necrotic spot virus (INSV) Tospovirus • Beet curly top virus (BCTV) Curtovirus Leafhopper *Whitefly-transmitted geminiviruses in peppers are not present in -

2020 Taxonomic Update for Phylum Negarnaviricota (Riboviria: Orthornavirae), Including the Large Orders Bunyavirales and Mononegavirales
Archives of Virology https://doi.org/10.1007/s00705-020-04731-2 VIROLOGY DIVISION NEWS 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales Jens H. Kuhn1 · Scott Adkins2 · Daniela Alioto3 · Sergey V. Alkhovsky4 · Gaya K. Amarasinghe5 · Simon J. Anthony6,7 · Tatjana Avšič‑Županc8 · María A. Ayllón9,10 · Justin Bahl11 · Anne Balkema‑Buschmann12 · Matthew J. Ballinger13 · Tomáš Bartonička14 · Christopher Basler15 · Sina Bavari16 · Martin Beer17 · Dennis A. Bente18 · Éric Bergeron19 · Brian H. Bird20 · Carol Blair21 · Kim R. Blasdell22 · Steven B. Bradfute23 · Rachel Breyta24 · Thomas Briese25 · Paul A. Brown26 · Ursula J. Buchholz27 · Michael J. Buchmeier28 · Alexander Bukreyev18,29 · Felicity Burt30 · Nihal Buzkan31 · Charles H. Calisher32 · Mengji Cao33,34 · Inmaculada Casas35 · John Chamberlain36 · Kartik Chandran37 · Rémi N. Charrel38 · Biao Chen39 · Michela Chiumenti40 · Il‑Ryong Choi41 · J. Christopher S. Clegg42 · Ian Crozier43 · John V. da Graça44 · Elena Dal Bó45 · Alberto M. R. Dávila46 · Juan Carlos de la Torre47 · Xavier de Lamballerie38 · Rik L. de Swart48 · Patrick L. Di Bello49 · Nicholas Di Paola50 · Francesco Di Serio40 · Ralf G. Dietzgen51 · Michele Digiaro52 · Valerian V. Dolja53 · Olga Dolnik54 · Michael A. Drebot55 · Jan Felix Drexler56 · Ralf Dürrwald57 · Lucie Dufkova58 · William G. Dundon59 · W. Paul Duprex60 · John M. Dye50 · Andrew J. Easton61 · Hideki Ebihara62 · Toufc Elbeaino63 · Koray Ergünay64 · Jorlan Fernandes195 · Anthony R. Fooks65 · Pierre B. H. Formenty66 · Leonie F. Forth17 · Ron A. M. Fouchier48 · Juliana Freitas‑Astúa67 · Selma Gago‑Zachert68,69 · George Fú Gāo70 · María Laura García71 · Adolfo García‑Sastre72 · Aura R. Garrison50 · Aiah Gbakima73 · Tracey Goldstein74 · Jean‑Paul J. Gonzalez75,76 · Anthony Grifths77 · Martin H. Groschup12 · Stephan Günther78 · Alexandro Guterres195 · Roy A. -
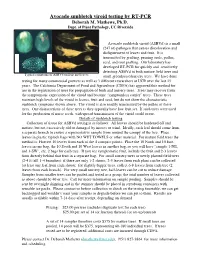
Avocado Sunblotch Viroid Testing by RT-PCR Deborah M
Avocado sunblotch viroid testing by RT-PCR Deborah M. Mathews, Ph.D. Dept. of Plant Pathology, UC Riverside Avocado sunblotch viroid (ASBVd) is a small (247 nt) pathogen that causes discoloration and disfigurement of leaves and fruit. It is transmitted by grafting, pruning tools, pollen, seed, and root grafting. Our laboratory has developed RT-PCR for quickly and sensitively detecting ASBVd in both mature field trees and Typical symptoms of ASBVd on fruit and leaves small greenhouse/nursery trees. We have done testing for many commercial growers as well as 3 different researchers at UCR over the last 15 years. The California Department of Food and Agriculture (CDFA) has approved this method for use in the registration of trees for propagation of buds and nursery trees. Trees may recover from the symptomatic expression of the viroid and become “symptomless carrier” trees. These trees maintain high levels of the viroid in leaves, fruit and seed, but do not show the characteristic sunblotch symptoms shown above. The viroid is also readily transmitted by the pollen of these trees. One characteristic of these trees is they typically have low fruit set. If such trees were used for the production of nurse seeds, widespread transmission of the viroid could occur. Details of sunblotch testing Collection of tissue for ASBVd testing is as follows: All leaves should be hardened off and mature, but not excessively old or damaged by insects or wind. Ideally, each leaf should come from a separate branch to ensure a representative sample from around the canopy of the tree. Place leaves in plastic ziplock bags with NO WET TOWELS or other material. -

Meadows Farms Nurseries Japanese Aucuba
Japanese Aucuba Aucuba japonica Height: 8 feet Spread: 8 feet Sunlight: Hardiness Zone: 6b Other Names: Spotted Laurel Japanese Aucuba foliage Description: Photo courtesy of NetPS Plant Finder An interesting evergreen shrub that solves the problem of the most shaded garden areas; female plants produce berries; ideal as a dense screen; drought tolerant once established Ornamental Features Japanese Aucuba has attractive yellow-spotted dark green foliage. The glossy pointy leaves are highly ornamental and remain dark green throughout the winter. Neither the flowers nor the fruit are ornamentally significant. Landscape Attributes Japanese Aucuba is a dense multi-stemmed evergreen shrub with a more or less rounded form. Its average texture blends into the landscape, but can be balanced by one or two finer or coarser trees or shrubs for an effective composition. This is a relatively low maintenance shrub, and can be pruned at anytime. It has no significant negative characteristics. Japanese Aucuba is recommended for the following landscape applications; - Accent - Mass Planting - Hedges/Screening - General Garden Use Planting & Growing Japanese Aucuba will grow to be about 8 feet tall at maturity, with a spread of 8 feet. It has a low canopy, and is suitable for planting under power lines. It grows at a fast rate, and under ideal conditions can be expected to live for approximately 20 years. This shrub does best in partial shade to shade. It does best in average to evenly moist conditions, but will not tolerate standing water. It is not particular as to soil pH, but grows best in rich soils. It is somewhat tolerant of urban pollution, and will benefit from being planted in a relatively sheltered location. -

Characterization and Genome Organization of New Luteoviruses and Nanoviruses Infecting Cool Season Food Legumes
Adane Abraham (Autor) Characterization and Genome Organization of New Luteoviruses and Nanoviruses Infecting Cool Season Food Legumes https://cuvillier.de/de/shop/publications/2549 Copyright: Cuvillier Verlag, Inhaberin Annette Jentzsch-Cuvillier, Nonnenstieg 8, 37075 Göttingen, Germany Telefon: +49 (0)551 54724-0, E-Mail: [email protected], Website: https://cuvillier.de CHAPTER 1 General Introduction Viruses and virus diseases of cool season food legumes Legume crops play a major role worldwide as source of human food, feed and also in crop rotation. Faba bean (Vicia faba L.), field pea (Pisum sativum L.), lentil (Lens culinaris Medik.), chickpea (Cicer arietinum L.), and grasspea (Lathyrus sativus L.), collectively re- ferred to as cool season food legumes (Summerfield et al. 1988) are of particular importance in developing countries of Asia, North and Northeast Africa where they provide a cheap source of seed protein for the predominantly poor population. Diseases including those caused by viruses are among the main constraints reducing their yield. Bos et al. (1988) listed some 44 viruses as naturally infecting faba bean, chickpea, field pea and lentil worldwide. Since then, a number of new viruses were described from these crops including Faba bean necrotic yellows virus (FBNYV) (Katul et al. 1993) and Chickpea chlorotic dwarf virus (CpCDV) (Horn et al. 1993), which are widespread and economically important. Most of the viruses of cool season food legumes are known to naturally infect more than one host within this group of crops (Bos et al. 1988, Brunt et al. 1996 and Makkouk et al. 2003a). Virus symptoms in cool season food legumes vary depending on the virus or its strain, host species or cultivar and the prevailing environmental conditions. -

Deciphering the Virome of Culex Vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan
viruses Article Deciphering the Virome of Culex vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan Astri Nur Faizah 1,2 , Daisuke Kobayashi 2,3, Haruhiko Isawa 2,*, Michael Amoa-Bosompem 2,4, Katsunori Murota 2,5, Yukiko Higa 2, Kyoko Futami 6, Satoshi Shimada 7, Kyeong Soon Kim 8, Kentaro Itokawa 9, Mamoru Watanabe 2, Yoshio Tsuda 2, Noboru Minakawa 6, Kozue Miura 1, Kazuhiro Hirayama 1,* and Kyoko Sawabe 2 1 Laboratory of Veterinary Public Health, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan; [email protected] (A.N.F.); [email protected] (K.M.) 2 Department of Medical Entomology, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo 162-8640, Japan; [email protected] (D.K.); [email protected] (M.A.-B.); k.murota@affrc.go.jp (K.M.); [email protected] (Y.H.); [email protected] (M.W.); [email protected] (Y.T.); [email protected] (K.S.) 3 Department of Research Promotion, Japan Agency for Medical Research and Development, 20F Yomiuri Shimbun Bldg. 1-7-1 Otemachi, Chiyoda-ku, Tokyo 100-0004, Japan 4 Department of Environmental Parasitology, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8510, Japan 5 Kyushu Research Station, National Institute of Animal Health, NARO, 2702 Chuzan, Kagoshima 891-0105, Japan 6 Department of Vector Ecology and Environment, Institute of Tropical Medicine, Nagasaki University, 1-12-4 Sakamoto, Nagasaki 852-8523, Japan; [email protected] -

Detection of Avocado Sunblotch Viroid and Estimation of Infection Among Accessions in the National Germplasm Collection for Avocado
Proc. Fla. State Hort. Soc. 109:235-237. 1996. DETECTION OF AVOCADO SUNBLOTCH VIROID AND ESTIMATION OF INFECTION AMONG ACCESSIONS IN THE NATIONAL GERMPLASM COLLECTION FOR AVOCADO Catherine M. Running and Raymond J. Schnell National Germplasm Repository U. S. Department of Agriculture Agricultural Research Service 13601 Old Cutler Road, Miami, FL 33158 David N. Kuhn Florida International University Dept. of Biological Sciences Miami, FL 33199 Additional index words. Persea Americana, RT-PCR, viroid, DNA, RNA, sequence variation, germplasm. ABSTRACT Reverse transcription-polymerase chain reaction (RTPCR) was used to determine the incidence of infection by avocado sunblotch viroid (ASBVd) in the germplasm collection at the National Germplasm Repository at Miami (NGR-Mia). Of the 429 avocado plants growing at the repository, 81 (18.9%) are infected with the viroid. The 429 plants represent 237 accessions. There are multiple plants of some accessions and for 42 accessions (17%) every plant is infected with the viroid. There was no apparent relationship between host race and the incidence of infection. Symptoms of sunblotch disease on avocado (Persea Americana Mill.) manifest as a general decline of tree vigor with sunken, yellow areas on the fruit that lessen its marketability. The disease was first described as infectious, rather than physiological, by Home and Parker (1931). The infectious agent has since been determined to be an RNA viroid known as Avocado Sunblotch Viroid (ASBVd) (Dale and Allen, 1979; Thomas and Mohammed, 1979). The viroid is transmitted by budding and grafting, including natural root grafting (Home etal, 1941; Whitsell, 1952), seed (Wallace and Drake, 1962), and pollen (Desjardins et al., 1979). -

Viral Diversity in Tree Species
Universidade de Brasília Instituto de Ciências Biológicas Departamento de Fitopatologia Programa de Pós-Graduação em Biologia Microbiana Doctoral Thesis Viral diversity in tree species FLÁVIA MILENE BARROS NERY Brasília - DF, 2020 FLÁVIA MILENE BARROS NERY Viral diversity in tree species Thesis presented to the University of Brasília as a partial requirement for obtaining the title of Doctor in Microbiology by the Post - Graduate Program in Microbiology. Advisor Dra. Rita de Cássia Pereira Carvalho Co-advisor Dr. Fernando Lucas Melo BRASÍLIA, DF - BRAZIL FICHA CATALOGRÁFICA NERY, F.M.B Viral diversity in tree species Flávia Milene Barros Nery Brasília, 2025 Pages number: 126 Doctoral Thesis - Programa de Pós-Graduação em Biologia Microbiana, Universidade de Brasília, DF. I - Virus, tree species, metagenomics, High-throughput sequencing II - Universidade de Brasília, PPBM/ IB III - Viral diversity in tree species A minha mãe Ruth Ao meu noivo Neil Dedico Agradecimentos A Deus, gratidão por tudo e por ter me dado uma família e amigos que me amam e me apoiam em todas as minhas escolhas. Minha mãe Ruth e meu noivo Neil por todo o apoio e cuidado durante os momentos mais difíceis que enfrentei durante minha jornada. Aos meus irmãos André, Diego e meu sobrinho Bruno Kawai, gratidão. Aos meus amigos de longa data Rafaelle, Evanessa, Chênia, Tati, Leo, Suzi, Camilets, Ricardito, Jorgito e Diego, saudade da nossa amizade e dos bons tempos. Amo vocês com todo o meu coração! Minha orientadora e grande amiga Profa Rita de Cássia Pereira Carvalho, a quem escolhi e fui escolhida para amar e fazer parte da família. -

Tically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 21 June 2021 doi:10.20944/preprints202106.0526.v1 Review Towards the forest virome: next-generation-sequencing dras- tically expands our understanding on virosphere in temperate forest ecosystems Artemis Rumbou 1,*, Eeva J. Vainio 2 and Carmen Büttner 1 1 Faculty of Life Sciences, Albrecht Daniel Thaer-Institute of Agricultural and Horticultural Sciences, Humboldt-Universität zu Berlin, Ber- lin, Germany; [email protected], [email protected] 2 Natural Resources Institute Finland, Latokartanonkaari 9, 00790, Helsinki, Finland; [email protected] * Correspondence: [email protected] Abstract: Forest health is dependent on the variability of microorganisms interacting with the host tree/holobiont. Symbiotic mi- crobiota and pathogens engage in a permanent interplay, which influences the host. Thanks to the development of NGS technol- ogies, a vast amount of genetic information on the virosphere of temperate forests has been gained the last seven years. To estimate the qualitative/quantitative impact of NGS in forest virology, we have summarized viruses affecting major tree/shrub species and their fungal associates, including fungal plant pathogens, mutualists and saprotrophs. The contribution of NGS methods is ex- tremely significant for forest virology. Reviewed data about viral presence in holobionts, allowed us to address the role of the virome in the holobionts. Genetic variation is a crucial aspect in hologenome, significantly reinforced by horizontal gene transfer among all interacting actors. Through virus-virus interplays synergistic or antagonistic relations may evolve, which may drasti- cally affect the health of the holobiont. Novel insights of these interplays may allow practical applications for forest plant protec- tion based on endophytes and mycovirus biocontrol agents. -

Molecular Identification and Characterization of Two Rubber Dandelion
Molecular Identification and Characterization of Two Rubber Dandelion Amalgaviruses Archives of Virology – Annotated Sequence Record Humberto Debat1*, Zinan Luo, Brian J. Iaffaldano, Xiaofeng Zhuang, Katrina Cornish 2 1 Instituto de Patología Vegetal, Centro de Investigaciones Agropecuarias, Instituto Nacional de Tecnología Agropecuaria (IPAVE-CIAP-INTA), 11 de setiembre 4755, Córdoba, Argentina, X5020ICA. 2 Department of Horticulture and Crop Science, The Ohio State University, Ohio Agricultural Research and Development Center, Williams Hall, 1680 Madison Avenue, Wooster, OH 44691. * Corresponding author: Humberto Debat, [email protected], Tel: +54 9 351 4973636, Fax: +54 9 351 4974330 Abstract The Amalgaviridae family is composed of persistent viruses that share the genome architecture of Totiviridae and gene evolutionary resemblance to Partitiviridae. A single Amalgavirus genus has been assigned to this family, harboring only four recognized species, corresponding to plant infecting viruses with dsRNA monopartite genomes of ca. 3.4 kb. Here, we present the genomic identification and characterization of two novel Amalgavirus detected in Rubber dandelion (Taraxacum kok-saghyz). The sequenced isolates presented a 3,409 and 3,413 nt long genome, harbouring two partially overlapping ORFs encoding a putative coat protein and an RNA-dependent RNA polymerase (RdRP). Multiple independent RNAseq data suggest that the identified viruses have a differential distribution and low relative RNA levels in infected plants. Virus presence was not associated with any apparent symptoms on the plant host. We propose the name rubber dandelion latent virus 1 & 2 to the detected Amalgavirus. Annotated sequence record Natural rubber is an essential material to the manufacture of 50,000 different rubber and latex products. -

A DISEASE OP AUCUBA JAPONICA THUNB. CONTENTS. Pag© I
A DISEASE OP AUCUBA JAPONICA THUNB. CONTENTS. Pag© I Introduction .................................. 1 II History of the Disease and Geographical Distribution ...... 1 III Economic Importance ........................... 2 IV Symptomatology ................................ 2 V Pathological Histology ........................ 6 (a) Stem ........................... 6 (b) Leaf ............................ 7 (c) Root ........................... 9 VI General Observations on the Diseased Tissues .. 10 VII Isolations from Typical Lesions .............. 10 VIII Inoculation Experiments ....................... 16 IX The Organism Associated with Aucuba Necrosis •. 19 (a) Morphology ..................... 19 (b) Staining Reactions ............. 20 (c) Cultural Characters ............ 20 (d) Physiological Properties ....... 22 (e) Technical Description .......... 50 X Action of Bacterial Toxins and Enzymes on Healthy Aucuba Tissue ..... 31 XI Inoculations with Bacterium-free Filtrate ..... 35 XII The Relation of Phomopsis Aucubae Trav. to Disease in Aucuba .... 34 XIII The Pathogenicity of Botrytis Cinerea Pers 37 XIV Experiments with Mixed Inocula ................ 46 XV Discussion .................................... 4?> XVI Summary ........................................ 63 References .................................... 65 Explanation of Plates .............. 66 ProQuest Number: 13905475 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a com plete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest ProQuest 13905475 Published by ProQuest LLC(2019). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States C ode Microform Edition © ProQuest LLC. ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106- 1346 A DISEASE OF AUCUBA JAPONICA THUNB.