Systemic Surfaceome Profiling Identifies Target Antigens for Immune-Based Therapy in Subtypes of Advanced Prostate Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

The Regulation of Interleukin 7 Receptor Alpha Internalization, Recycling and Degradation by IL-7
Universidade de Lisboa Faculdade de Medicina Unidade de Biologia do Cancro, Instituto de Medicina Molecular The regulation of Interleukin 7 receptor alpha internalization, recycling and degradation by IL-7 - Possible implications in T-cell homeostasis, migration and leukaemogenesis - Catarina Martins de Oliveira Henriques Doutoramento em Ciências Biomédicas For the degree of Doctor of Philosophy 2009 Universidade de Lisboa Faculdade de Medicina Unidade de Biologia do Cancro, Instituto de Medicina Molecular The regulation of Interleukin 7 receptor alpha internalization, recycling and degradation by IL-7 - Possible implications in T-cell homeostasis, migration and leukaemogenesis - Catarina Martins de Oliveira Henriques (Recipient of a scholarship- SFRH7BD/21940/2005 from Fundação para a Ciência e Tecnologia) Tese orientada pelo Doutor João T. Barata, Prof Doutor.Gerard Graham e Prof. Doutora Leonor Parreira Doutoramento em Ciências Biomédicas, especialidade em Ciências Biopatológicas For the degree of Doctor of Philosophy 2009 As opiniões expressas são da exclusiva responsabilidade do seu autor A impressão desta dissertação foi aprovada pela Comissão Coordenadora do Conselho Científico da Faculdade de Medicina de Lisboa em reunião de 13 de Outubro de 2009. Para a Prof. Filomena Mota. Sem o seu apoio e inspiração há muitos anos atrás, eu não seria hoje uma bióloga nem esta tese teria alguma vez existido… Table of Contents Table of contents……………………………………………………….………………….................. i Aknowledgements……………………………………………………………………………………………. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

The Immunomodulatory CEA Cell Adhesion Molecule 6 (CEACAM6/Cd66c) Is a Candidate Receptor for the Influenza a Virus
bioRxiv preprint doi: https://doi.org/10.1101/104026; this version posted January 30, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 The immunomodulatory CEA cell adhesion molecule 6 (CEACAM6/CD66c) is a 2 candidate receptor for the influenza A virus 3 Shah Kamranur Rahmana *, Mairaj Ahmed Ansarib, Pratibha Gaurc, Imtiyaz Ahmada, 4 Chandrani Chakravartya,d, Dileep Kumar Vermaa, Sanjay Chhibbere, Naila Nehalf, 5 Shanmugaapriya Sellathanbyd, Dagmar Wirthc, Gulam Warisb and Sunil K. Lala,g # 6 7 Virology Group, International Centre for Genetic Engineering & Biotechnology, New Delhi, 8 Indiaa. 9 Department of Microbiology and Immunology, H. M. Bligh Cancer Research Laboratories, 10 Rosalind Franklin University of Medicine and Science, Chicago Medical School, North 11 Chicago, Illinois, USAb. 12 Helmholtz Centre for Infection Research, Braunschweig, Germanyc. 13 Department of Biomedical Science, Bharathidasan University, Trichy, Indiad. 14 Microbiology Department, Panjab University, Chandigarh, Indiae. 15 Career Institute of Medical & Dental Sciences and Hospital, Lucknow, Indiaf. 16 School of Science, Monash University, Selangor DE, Malaysiag. 17 18 Running Head: Protein receptor for Influenza A Virus 19 20 # Corresponding author: Professor of Microbiology, School of Science, Monash University, 21 47500 Bandar Sunway, Selangor DE, Malaysia. 22 Email: [email protected]; Telephone: (+603) 551 59606 23 24 * Current address: Department of Pathogen Molecular Biology, London School of Hygiene & 25 Tropical Medicine, Keppel Street, London WC1E 7HT, United Kingdom. 26 1 bioRxiv preprint doi: https://doi.org/10.1101/104026; this version posted January 30, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Pan-Cancer Transcriptome Analysis Reveals a Gene Expression
Modern Pathology (2016) 29, 546–556 546 © 2016 USCAP, Inc All rights reserved 0893-3952/16 $32.00 Pan-cancer transcriptome analysis reveals a gene expression signature for the identification of tumor tissue origin Qinghua Xu1,6, Jinying Chen1,6, Shujuan Ni2,3,4,6, Cong Tan2,3,4,6, Midie Xu2,3,4, Lei Dong2,3,4, Lin Yuan5, Qifeng Wang2,3,4 and Xiang Du2,3,4 1Canhelp Genomics, Hangzhou, Zhejiang, China; 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China; 3Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China; 4Institute of Pathology, Fudan University, Shanghai, China and 5Pathology Center, Shanghai General Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China Carcinoma of unknown primary, wherein metastatic disease is present without an identifiable primary site, accounts for ~ 3–5% of all cancer diagnoses. Despite the development of multiple diagnostic workups, the success rate of primary site identification remains low. Determining the origin of tumor tissue is, thus, an important clinical application of molecular diagnostics. Previous studies have paved the way for gene expression-based tumor type classification. In this study, we have established a comprehensive database integrating microarray- and sequencing-based gene expression profiles of 16 674 tumor samples covering 22 common human tumor types. From this pan-cancer transcriptome database, we identified a 154-gene expression signature that discriminated the origin of tumor tissue with an overall leave-one-out cross-validation accuracy of 96.5%. The 154-gene expression signature was first validated on an independent test set consisting of 9626 primary tumors, of which 97.1% of cases were correctly classified. -
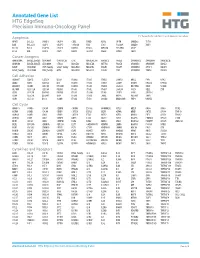
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

Dnmt3b Overexpression Contributes to a Hypermethylator Phenotype in Human Breast Cancer Cell Lines
Molecular Cancer BioMed Central Research Open Access DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines J Devon Roll1,3, Ashley G Rivenbark2,3, Wendell D Jones4 and William B Coleman*1,3 Address: 1Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC 27599, USA, 2Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA, 3UNC Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA and 4Expression Analysis, 2605 Meridian Parkway, Durham, NC 27713, USA Email: J Devon Roll - [email protected]; Ashley G Rivenbark - [email protected]; Wendell D Jones - [email protected]; William B Coleman* - [email protected] * Corresponding author Published: 25 January 2008 Received: 5 November 2007 Accepted: 25 January 2008 Molecular Cancer 2008, 7:15 doi:10.1186/1476-4598-7-15 This article is available from: http://www.molecular-cancer.com/content/7/1/15 © 2008 Roll et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: DNA hypermethylation events and other epimutations occur in many neoplasms, producing gene expression changes that contribute to neoplastic transformation, tumorigenesis, and tumor behavior. Some human cancers exhibit a hypermethylator phenotype, characterized by concurrent DNA methylation-dependent silencing of multiple genes. To determine if a hypermethylation defect occurs in breast cancer, the expression profile and promoter methylation status of methylation-sensitive genes were evaluated among breast cancer cell lines. -

Departments of 3Oncology and 4Pathology, St. Jude
UNIVERSAL MONITORING OF MINIMAL RESIDUAL DISEASE IN ACUTE MYELOID LEUKEMIA Elaine Coustan-Smith,1 Guangchun Song,4 Sheila Shurtleff,4 Allen Eng-Juh Yeoh,1,2 Chng Wee Joo,2 Siew Peng Chen,1 Jeffrey E. Rubnitz,3,5 Ching-Hon Pui,3,4,5 James R. Downing,4,5 Dario Campana1,2 1Department of Pediatrics and 2National University Cancer Institute Singapore, National University of Singapore, Singapore; Departments of 3Oncology and 4Pathology, St. Jude Children's Research Hospital, Memphis, TN; and 5University of Tennessee Health Science Center, Memphis, TN Supplemental Material (Supplemental Tables S3-S6; Supplemental Figures S1-S6) 1 Table S3. Genes overexpressed in AML “stem cells” according to previous studies and their overexpression in AML according to the present analysis Gene overexpressed in AML Gene overexpressed in AML AML cases with stem cells according to Saito et stem cells according to Kikushige overexpression in this al.(1) et al.(2) study (%)a WT1 84.7 CD32 CD32 76.4 DOK2 70.1 CD96 66.9 HCK 65.6 CD86 CD86 64.3 CD44 64.3 CD93 56.7 ITGB2, CD18 ITGB2, CD18 52.9 CSF1R, CD115 51.0 IL2RA, CD25 IL2RA, CD25 47.8 LY86 43.3 IL7R, CD127 42.0 CD99 42.0 IL17R 39.5 CD97 CD97 37.6 CD33 CD33 36.9 CD9 36.9 CD1C 35.7 AK5 33.1 BIK 31.2 CD47 30.6 TNFRSF4, CD134 29.3 CD84 29.3 IL2RG, CD132 27.4 ITGB7 27.4 CEACAM6, CD66c 26.8 FLT3 25.5 CD180 <25 CTSC <25 PDE9A <25 CD24 <25 CD36 <25 CD123 <25 ITGAE <25 2 LRG1 Not on HG-U133A array SUCNR1 Not on HG-U133A array TNFSF13B, CD257 Not on HG-U133A array CD366, HAVCR2, TIM-3 Not on HG-U133A array CD371, CLEC12A, CLL-1 Not on HG-U133A array aGene expression was studied by HG-U133A oligonucleotide microarrays in157 AML samples and 7 samples of normal CD34+ myeloid progenitors. -

Soluble Carcinoembryonic Antigen Activates Endothelial Cells and Tumor Angiogenesis
Author Manuscript Published OnlineFirst on October 11, 2013; DOI: 10.1158/0008-5472.CAN-13-0123 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Soluble carcinoembryonic antigen activates endothelial cells and tumor angiogenesis Kira H. Bramswig,1,5 Marina Poettler,1,5 Matthias Unseld,1 Friedrich Wrba,2 Pavel Uhrin,3 Wolfgang Zimmermann,4 Christoph C. Zielinski,1 Gerald W. Prager1,* 1 Clinical Division of Oncology, Department of Medicine I and Cancer Center, Medical University of Vienna, Austria 2 Institute of Clinical Pathology, Medical University of Vienna, Austria 3 Department of Vascular Biology and Thrombosis Research, Centre for Bio-Molecular Medicine and Pharmacology, Medical University of Vienna, Austria 4 Tumor Immunology Laboratory, LIFE-Center, Klinikum Grosshadern, Ludwig-Maximilians- University Munich 5 These authors contributed equally to this work *Correspondence: Dr. Gerald Prager Waehringer Guertel 18-20, A-1090 Vienna, Austria Tel: 0043-1-40400-4450 Fax: 0043-1-40400-4451 [email protected] Running Title: CEA affects angiogenesis Conflicts of interest: None Abstract: 183 words Text-Body without References: 4998 words Keywords: carcinoembryonic antigen, endothelial cells, integrins, tumor-angiogenesis 1 Downloaded from cancerres.aacrjournals.org on September 25, 2021. © 2013 American Association for Cancer Research. Author Manuscript Published OnlineFirst on October 11, 2013; DOI: 10.1158/0008-5472.CAN-13-0123 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Abstract Carcinoembryonic antigen (CEA, CD66e, CEACAM-5) is a cell-surface bound glycoprotein overexpressed and released by many solid tumors that has an autocrine function in cancer cell survival and differentiation. -

In Pancreatic Ductal Adenocarcinoma (PDA): an Integrative Analysis of a Novel Therapeutic Target
Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in Pancreatic Ductal Adenocarcinoma (PDA): An integrative analysis of a novel therapeutic target Item Type Article Authors Pandey, Ritu; Zhou, Muhan; Islam, Shariful; Chen, Baowei; Barker, Natalie K; Langlais, Paul; Srivastava, Anup; Luo, Moulun; Cooke, Laurence S; Weterings, Eric; Mahadevan, Daruka Citation Pandey, R., Zhou, M., Islam, S. et al. Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in Pancreatic Ductal Adenocarcinoma (PDA): An integrative analysis of a novel therapeutic target. Sci Rep 9, 18347 (2019). https:// doi.org/10.1038/s41598-019-54545-9 DOI 10.1038/s41598-019-54545-9 Publisher NATURE PUBLISHING GROUP Journal SCIENTIFIC REPORTS Rights Copyright © The Author(s) 2019. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License. Download date 25/09/2021 09:11:41 Item License https://creativecommons.org/licenses/by/4.0/ Version Final published version Link to Item http://hdl.handle.net/10150/636770 www.nature.com/scientificreports OPEN Carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) in Pancreatic Ductal Adenocarcinoma (PDA): An integrative analysis of a novel therapeutic target Ritu Pandey1,2*, Muhan Zhou1, Shariful Islam1, Baowei Chen1, Natalie K Barker3, Paul Langlais3, Anup Srivastava3, Moulun Luo3, Laurence S. Cooke1, Eric Weterings1,3,4 & Daruka Mahadevan1,3* We investigated biomarker CEACAM6, a highly abundant cell surface adhesion receptor that modulates the extracellular matrix (ECM) in pancreatic ductal adenocarcinoma (PDA). The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) RNA-Seq data from PDA patients were analyzed for CEACAM6 expression and evaluated for overall survival, association, enrichment and correlations. -

A Potential Role of IL-6/IL-6R in the Development and Management of Colon Cancer
membranes Review A Potential Role of IL-6/IL-6R in the Development and Management of Colon Cancer Mimmo Turano 1 , Francesca Cammarota 2,3, Francesca Duraturo 2,3, Paola Izzo 2,3 and Marina De Rosa 2,3,* 1 Department of Biology, University of Naples Federico II, 80126 Naples, Italy; [email protected] 2 Department of Molecular Medicine and Medical Biotechnology, University of Naples Federico II, 80131 Naples, Italy; [email protected] (F.C.); [email protected] (F.D.); [email protected] (P.I.) 3 Ceinge Biotecnologie Avanzate, 80131 Naples, Italy * Correspondence: [email protected] Abstract: Colorectal cancer (CRC) is the third most frequent cancer worldwide and the second greatest cause of cancer deaths. About 75% of all CRCs are sporadic cancers and arise following somatic mutations, while about 10% are hereditary cancers caused by germline mutations in specific genes. Several factors, such as growth factors, cytokines, and genetic or epigenetic alterations in specific oncogenes or tumor-suppressor genes, play a role during the adenoma–carcinoma sequence. Recent studies have reported an increase in interleukin-6 (IL-6) and soluble interleukin-6 receptor (sIL-6R) levels in the sera of patients affected by colon cancer that correlate with the tumor size, suggesting a potential role for IL-6 in colon cancer progression. IL-6 is a pleiotropic cytokine showing both pro- and anti-inflammatory roles. Two different types of IL-6 signaling are known. Classic IL-6 signaling involves the binding of IL-6 to its membrane receptor on the surfaces of target cells; alternatively, IL-6 binds to sIL-6R in a process called IL-6 trans-signaling.