Novel Gene Signatures Predictive of Patient Recurrence-Free Survival and Castration Resistance in Prostate Cancer Jun A, Baotong Zhang, Zhiqian Zhang, Jin-Tang Dong
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Contribution of MSMB Promoter Region Gene Polymorphism to Early-Onset Prostate Cancer Risk in Mexican Males
www.oncotarget.com Oncotarget, 2019, Vol. 10, (No. 7), pp: 738-748 Research Paper Contribution of MSMB promoter region gene polymorphism to early-onset prostate cancer risk in Mexican males Silvia Juliana Trujillo-Cáceres1, Luisa Torres-Sánchez1, Ana I. Burguete-García2, Yaneth Citlalli Orbe Orihuela2, Ruth Argelia Vázquez-Salas3, Esmeralda Álvarez- Topete4 and Rocío Gómez4 1Centro de Investigación en Salud Poblacional, Instituto Nacional de Salud Pública (INSP), Cuernavaca, Morelos, Mexico 2Centro de Investigación en Enfermedades Infecciosas, INSP, Cuernavaca, Morelos, Mexico 3Conacyt-Centro de Investigación en Salud Poblacional, INSP, Cuernavaca, Morelos, Mexico 4Departamento de Toxicología, Cinvestav-IPN, Mexico City, Mexico Correspondence to: Luisa Torres-Sánchez, email: [email protected] Keywords: genetic polymorphisms; MSMB; prostate cancer; rs10993994; sexually transmitted diseases Received: July 13, 2018 Accepted: December 16, 2018 Published: January 22, 2019 Copyright: Trujillo-Cáceres et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Sexually transmitted infections and its contribution to prostate cancer (PC) development have been relevant in different populations. MSMB gene polymorphism (rs10993994) has exhibited an association both with PC as well as the susceptibility to sexually transmitted infections. Hitherto, these conditions have been not studied in Mexico yet, neither if sexually transmitted infections could modify the MSMB and PC association. Herein, socio-demographic features, sexually transmitted infections records, the reproductive backgrounds, and the genetic characterisation were analysed in 322 incident PC cases and 628 population healthy controls from Mexico City. -

DF6720-MSMB Antibody
Affinity Biosciences website:www.affbiotech.com order:[email protected] MSMB Antibody Cat.#: DF6720 Concn.: 1mg/ml Mol.Wt.: 13kDa Size: 50ul,100ul,200ul Source: Rabbit Clonality: Polyclonal Application: WB 1:500-1:2000, IHC 1:50-1:200, ELISA(peptide) 1:20000-1:40000 *The optimal dilutions should be determined by the end user. Reactivity: Human,Mouse Purification: The antiserum was purified by peptide affinity chromatography using SulfoLink™ Coupling Resin (Thermo Fisher Scientific). Specificity: MSMB Antibody detects endogenous levels of total MSMB. Immunogen: A synthesized peptide derived from human MSMB, corresponding to a region within the internal amino acids. Uniprot: P08118 Description: The protein encoded by this gene is a member of the immunoglobulin binding factor family. It is synthesized by the epithelial cells of the prostate gland and secreted into the seminal plasma. This protein has inhibin-like activity. It may have a role as an autocrine paracrine factor in uterine, breast and other female reproductive tissues. The expression of the encoded protein is found to be decreased in prostate cancer. Two alternatively spliced transcript variants encoding different isoforms are described for this gene. The use of alternate polyadenylation sites has been found for this gene. Storage Condition and Rabbit IgG in phosphate buffered saline , pH 7.4, 150mM Buffer: NaCl, 0.02% sodium azide and 50% glycerol.Store at -20 °C.Stable for 12 months from date of receipt. Western blot analysis of HepG2 cell lysates, using MSMB Antibody. The lane on the left was treated with the antigen- specific peptide. 1 / 2 Affinity Biosciences website:www.affbiotech.com order:[email protected] DF6720 at 1/100 staining Mouse liver tissue by IHC-P. -

Supplementary Tables
Supp Table 1. Patient characteristics of samples used for colony assays, xenografts and intracellular colony assays Sr.No Age Sex Diagnosis Blast% Cytogenetics Mutations (IPSS) (AF%) Patient samples used for colony assays 1. 68 F Low risk 1% N/A None MDS 2. 78 M RAEB-1 7% N/A DNMT3A 3. 66 F High risk 12.2% 5q-,7q-,+11, DNMT3A MDS 20q- (28%) TP53 (58%) 4. 61 M High risk 6.2% Monosomy 7 ASXL1 (36%) MDS EZH2 (77%) RUNX1(18%) 5. 63 M Low risk Normal ETV6 (34%), MDS KRAS(15%), RUNX1 (40%), SRSF2 (43%), ZRSR2 (86%) 6. 62 F RAEB-2 Del 5q TP53 (7%) 7. 76 F t-MDS <1% Normal TET2(10%) 8. 80 M Low risk 5% Normal SF3B1(21%) MDS TET2(8%) ZRZR2(62%) 9. 74 F MPN 5% JAK2V617F+ 10. 76 M Low risk <1% Deletion Y None MDS 11. 84 M Low risk N/A N/A N/A MDS 12. 86 M Int-2 MDS 4-8% Normal U2AF1 (43%) blasts CBL (15%) 13. 64 M low risk 1-3% 20q deletion ASXL1 (17%) MDS SETBP1(17%) U2AF1(15%) 14. 81 F Low risk <1% Normal None MDS Patient samples used for PDX 15. 87 F Int-2 risk 1.2% Complex SETBP1 (38%) MDS cytogenetics, del 7, dup 11, del 13q 16. 59 F High-risk 7-10% Complex NRAS (12%), MDS cytogenetics RUNX1 (20%), (-5q31, -7q31, SRSF2 (22%), trisomy 8, del STAG2 (17%) 11q23 17. 79 F Int-2 risk 5-8% None MDS 18. 67 M MPN 6.6% Normal CALR (51%) IDH1(47%) PDGFRB (47%) Patient samples used for intracellular ASO uptake 19. -

NICU Gene List Generator.Xlsx
Neonatal Crisis Sequencing Panel Gene List Genes: A2ML1 - B3GLCT A2ML1 ADAMTS9 ALG1 ARHGEF15 AAAS ADAMTSL2 ALG11 ARHGEF9 AARS1 ADAR ALG12 ARID1A AARS2 ADARB1 ALG13 ARID1B ABAT ADCY6 ALG14 ARID2 ABCA12 ADD3 ALG2 ARL13B ABCA3 ADGRG1 ALG3 ARL6 ABCA4 ADGRV1 ALG6 ARMC9 ABCB11 ADK ALG8 ARPC1B ABCB4 ADNP ALG9 ARSA ABCC6 ADPRS ALK ARSL ABCC8 ADSL ALMS1 ARX ABCC9 AEBP1 ALOX12B ASAH1 ABCD1 AFF3 ALOXE3 ASCC1 ABCD3 AFF4 ALPK3 ASH1L ABCD4 AFG3L2 ALPL ASL ABHD5 AGA ALS2 ASNS ACAD8 AGK ALX3 ASPA ACAD9 AGL ALX4 ASPM ACADM AGPS AMELX ASS1 ACADS AGRN AMER1 ASXL1 ACADSB AGT AMH ASXL3 ACADVL AGTPBP1 AMHR2 ATAD1 ACAN AGTR1 AMN ATL1 ACAT1 AGXT AMPD2 ATM ACE AHCY AMT ATP1A1 ACO2 AHDC1 ANK1 ATP1A2 ACOX1 AHI1 ANK2 ATP1A3 ACP5 AIFM1 ANKH ATP2A1 ACSF3 AIMP1 ANKLE2 ATP5F1A ACTA1 AIMP2 ANKRD11 ATP5F1D ACTA2 AIRE ANKRD26 ATP5F1E ACTB AKAP9 ANTXR2 ATP6V0A2 ACTC1 AKR1D1 AP1S2 ATP6V1B1 ACTG1 AKT2 AP2S1 ATP7A ACTG2 AKT3 AP3B1 ATP8A2 ACTL6B ALAS2 AP3B2 ATP8B1 ACTN1 ALB AP4B1 ATPAF2 ACTN2 ALDH18A1 AP4M1 ATR ACTN4 ALDH1A3 AP4S1 ATRX ACVR1 ALDH3A2 APC AUH ACVRL1 ALDH4A1 APTX AVPR2 ACY1 ALDH5A1 AR B3GALNT2 ADA ALDH6A1 ARFGEF2 B3GALT6 ADAMTS13 ALDH7A1 ARG1 B3GAT3 ADAMTS2 ALDOB ARHGAP31 B3GLCT Updated: 03/15/2021; v.3.6 1 Neonatal Crisis Sequencing Panel Gene List Genes: B4GALT1 - COL11A2 B4GALT1 C1QBP CD3G CHKB B4GALT7 C3 CD40LG CHMP1A B4GAT1 CA2 CD59 CHRNA1 B9D1 CA5A CD70 CHRNB1 B9D2 CACNA1A CD96 CHRND BAAT CACNA1C CDAN1 CHRNE BBIP1 CACNA1D CDC42 CHRNG BBS1 CACNA1E CDH1 CHST14 BBS10 CACNA1F CDH2 CHST3 BBS12 CACNA1G CDK10 CHUK BBS2 CACNA2D2 CDK13 CILK1 BBS4 CACNB2 CDK5RAP2 -

Association of Imputed Prostate Cancer Transcriptome with Disease Risk Reveals Novel Mechanisms
Corrected: Author Correction ARTICLE https://doi.org/10.1038/s41467-019-10808-7 OPEN Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms Nima C. Emami1,2, Linda Kachuri2, Travis J. Meyers2, Rajdeep Das3,4, Joshua D. Hoffman2, Thomas J. Hoffmann 2,5, Donglei Hu 5,6,7, Jun Shan8, Felix Y. Feng3,4,7, Elad Ziv5,6,7, Stephen K. Van Den Eeden 3,8 & John S. Witte1,2,3,5,7 1234567890():,; Here we train cis-regulatory models of prostate tissue gene expression and impute expression transcriptome-wide for 233,955 European ancestry men (14,616 prostate cancer (PrCa) cases, 219,339 controls) from two large cohorts. Among 12,014 genes evaluated in the UK Biobank, we identify 38 associated with PrCa, many replicating in the Kaiser Permanente RPGEH. We report the association of elevated TMPRSS2 expression with increased PrCa risk (independent of a previously-reported risk variant) and with increased tumoral expression of the TMPRSS2:ERG fusion-oncogene in The Cancer Genome Atlas, suggesting a novel germline-somatic interaction mechanism. Three novel genes, HOXA4, KLK1, and TIMM23, additionally replicate in the RPGEH cohort. Furthermore, 4 genes, MSMB, NCOA4, PCAT1, and PPP1R14A, are associated with PrCa in a trans-ethnic meta-analysis (N = 9117). Many genes exhibit evidence for allele-specific transcriptional activation by PrCa master-regulators (including androgen receptor) in Position Weight Matrix, Chip-Seq, and Hi-C experimental data, suggesting common regulatory mechanisms for the associated genes. 1 Program in Biological and Medical Informatics, University of California San Francisco, San Francisco, CA 94158, USA. -

Early Growth Response 1 Regulates Hematopoietic Support and Proliferation in Human Primary Bone Marrow Stromal Cells
Hematopoiesis SUPPLEMENTARY APPENDIX Early growth response 1 regulates hematopoietic support and proliferation in human primary bone marrow stromal cells Hongzhe Li, 1,2 Hooi-Ching Lim, 1,2 Dimitra Zacharaki, 1,2 Xiaojie Xian, 2,3 Keane J.G. Kenswil, 4 Sandro Bräunig, 1,2 Marc H.G.P. Raaijmakers, 4 Niels-Bjarne Woods, 2,3 Jenny Hansson, 1,2 and Stefan Scheding 1,2,5 1Division of Molecular Hematology, Department of Laboratory Medicine, Lund University, Lund, Sweden; 2Lund Stem Cell Center, Depart - ment of Laboratory Medicine, Lund University, Lund, Sweden; 3Division of Molecular Medicine and Gene Therapy, Department of Labora - tory Medicine, Lund University, Lund, Sweden; 4Department of Hematology, Erasmus MC Cancer Institute, Rotterdam, the Netherlands and 5Department of Hematology, Skåne University Hospital Lund, Skåne, Sweden ©2020 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2019.216648 Received: January 14, 2019. Accepted: July 19, 2019. Pre-published: August 1, 2019. Correspondence: STEFAN SCHEDING - [email protected] Li et al.: Supplemental data 1. Supplemental Materials and Methods BM-MNC isolation Bone marrow mononuclear cells (BM-MNC) from BM aspiration samples were isolated by density gradient centrifugation (LSM 1077 Lymphocyte, PAA, Pasching, Austria) either with or without prior incubation with RosetteSep Human Mesenchymal Stem Cell Enrichment Cocktail (STEMCELL Technologies, Vancouver, Canada) for lineage depletion (CD3, CD14, CD19, CD38, CD66b, glycophorin A). BM-MNCs from fetal long bones and adult hip bones were isolated as reported previously 1 by gently crushing bones (femora, tibiae, fibulae, humeri, radii and ulna) in PBS+0.5% FCS subsequent passing of the cell suspension through a 40-µm filter. -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Alterations in LMTK2, MSMB and HNF1B Gene Expression Are Associated with the Development of Prostate Cancer BMC Cancer 2010, 10:315
Harries et al. BMC Cancer 2010, 10:315 http://www.biomedcentral.com/1471-2407/10/315 RESEARCH ARTICLE Open Access AlterationsResearch article in LMTK2, MSMB and HNF1B gene expression are associated with the development of prostate cancer Lorna W Harries*1, John RB Perry2, Paul McCullagh3 and Malcolm Crundwell4 Abstract Background: Genome wide association studies (GWAS) have identified several genetic variants that are associated with prostate cancer. Most of these variants, like other GWAS association signals, are located in non-coding regions of potential candidate genes, and thus could act at the level of the mRNA transcript. Methods: We measured the expression and isoform usage of seven prostate cancer candidate genes in benign and malignant prostate by real-time PCR, and correlated these factors with cancer status and genotype at the GWAS risk variants. Results: We determined that levels of LMTK2 transcripts in prostate adenocarcinomas were only 32% of those in benign tissues (p = 3.2 × 10-7), and that an independent effect of genotype at variant rs6465657 on LMTK2 expression in benign (n = 39) and malignant tissues (n = 21) was also evident (P = 0.002). We also identified that whilst HNF1B(C) and MSMB2 comprised the predominant isoforms in benign tissues (90% and 98% of total HNF1B or MSMB expression), HNF1B(B) and MSMB1 were predominant in malignant tissue (95% and 96% of total HNF1B or MSMB expression; P = 1.7 × 10-7 and 4 × 10-4 respectively), indicating major shifts in isoform usage. Conclusions: Our results indicate that the amount or nature of mRNA transcripts expressed from the LMTK2, HNF1B and MSMB candidate genes is altered in prostate cancer, and provides further evidence for a role for these genes in this disorder. -

LITAF and TNFSF15, Two Downstream Targets of AMPK, Exert Inhibitory Effects on Tumor Growth
Oncogene (2011) 30, 1892–1900 & 2011 Macmillan Publishers Limited All rights reserved 0950-9232/11 www.nature.com/onc ORIGINAL ARTICLE LITAF and TNFSF15, two downstream targets of AMPK, exert inhibitory effects on tumor growth J Zhou1, Z Yang2, T Tsuji3, J Gong1, J Xie1, C Chen3,WLi1, S Amar4 and Z Luo1 1Department of Biochemistry, Boston University School of Medicine, Boston, MA, USA; 2Department of Biochemistry and Molecular Biology, College of Life Sciences, Shaanxi Normal University, Xi’an, PR China; 3Department of Radiation Oncology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, USA and 4Department of Periodontology and Oral Biology, Boston University School of Dental Medicine, Boston, MA, USA Lipopolysaccharide (LPS)-induced tumor necrosis factor Keywords: AMPK; LITAF; TNFSF15; p53; tumor (TNF) a factor (LITAF) is a multiple functional molecule suppressor; tumorigenesis whose sequence is identical to the small integral membrane protein of the lysosome/late endosome. LITAF was initially identified as a transcription factor that activates transcription of proinflammatory cytokine in Introduction macrophages in response to LPS. Mutations of the LITAF gene are associated with a genetic disease, called Lipopolysaccharide (LPS)-induced tumor necrosis fac- Charcot–Marie–Tooth syndrome. Recently, we have tor (TNF) a factor (LITAF) was initially characterized reported that mRNA levels of LITAF and TNF super- as a transcription factor that activates transcription of family member 15 (TNFSF15) are upregulated by 50 cytokines such as TNFa, interleukin 6, soluble TNF- adenosine monophosphate (AMP)-activated protein ki- receptor II (sTNF-RII) and chemokine interleukin-8 in nase (AMPK). The present study further assesses their macrophages in response to LPS (Myokai et al., 1999; biological functions. -

A Single-Cell Transcriptome Atlas of the Mouse Glomerulus
RAPID COMMUNICATION www.jasn.org A Single-Cell Transcriptome Atlas of the Mouse Glomerulus Nikos Karaiskos,1 Mahdieh Rahmatollahi,2 Anastasiya Boltengagen,1 Haiyue Liu,1 Martin Hoehne ,2 Markus Rinschen,2,3 Bernhard Schermer,2,4,5 Thomas Benzing,2,4,5 Nikolaus Rajewsky,1 Christine Kocks ,1 Martin Kann,2 and Roman-Ulrich Müller 2,4,5 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background Three different cell types constitute the glomerular filter: mesangial depending on cell location relative to the cells, endothelial cells, and podocytes. However, to what extent cellular heteroge- glomerular vascular pole.3 Because BP ad- neity exists within healthy glomerular cell populations remains unknown. aptation and mechanoadaptation of glo- merular cells are key determinants of kidney Methods We used nanodroplet-based highly parallel transcriptional profiling to function and dysregulated in kidney disease, characterize the cellular content of purified wild-type mouse glomeruli. we tested whether glomerular cell type sub- Results Unsupervised clustering of nearly 13,000 single-cell transcriptomes identi- sets can be identified by single-cell RNA fied the three known glomerular cell types. We provide a comprehensive online sequencing in wild-type glomeruli. This atlas of gene expression in glomerular cells that can be queried and visualized using technique allows for high-throughput tran- an interactive and freely available database. Novel marker genes for all glomerular scriptome profiling of individual cells and is cell types were identified and supported by immunohistochemistry images particularly suitable for identifying novel obtained from the Human Protein Atlas. -

Overview of Research on Fusion Genes in Prostate Cancer
2011 Review Article Overview of research on fusion genes in prostate cancer Chunjiao Song1,2, Huan Chen3 1Medical Research Center, Shaoxing People’s Hospital, Shaoxing University School of Medicine, Shaoxing 312000, China; 2Shaoxing Hospital, Zhejiang University School of Medicine, Shaoxing 312000, China; 3Key Laboratory of Microorganism Technology and Bioinformatics Research of Zhejiang Province, Zhejiang Institute of Microbiology, Hangzhou 310000, China Contributions: (I) Conception and design: C Song; (II) Administrative support: Shaoxing Municipal Health and Family Planning Science and Technology Innovation Project (2017CX004) and Shaoxing Public Welfare Applied Research Project (2018C30058); (III) Provision of study materials or patients: None; (IV) Collection and assembly of data: C Song; (V) Data analysis and interpretation: H Chen; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Chunjiao Song. No. 568 Zhongxing Bei Road, Shaoxing 312000, China. Email: [email protected]. Abstract: Fusion genes are known to drive and promote carcinogenesis and cancer progression. In recent years, the rapid development of biotechnologies has led to the discovery of a large number of fusion genes in prostate cancer specimens. To further investigate them, we summarized the fusion genes. We searched related articles in PubMed, CNKI (Chinese National Knowledge Infrastructure) and other databases, and the data of 92 literatures were summarized after preliminary screening. In this review, we summarized approximated 400 fusion genes since the first specific fusion TMPRSS2-ERG was discovered in prostate cancer in 2005. Some of these are prostate cancer specific, some are high-frequency in the prostate cancer of a certain ethnic group. This is a summary of scientific research in related fields and suggests that some fusion genes may become biomarkers or the targets for individualized therapies. -
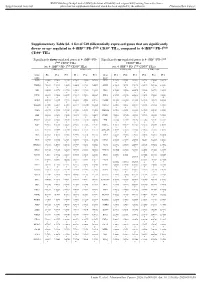
Supplementary Table S6. a List of 520 Differentially Expressed Genes
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Immunother Cancer Supplementary Table S6. A list of 520 differentially expressed genes that are significantly down- or up- regulated in 4-1BBpos PD-1high CD39+ TILs, compared to 4-1BBneg PD-1high CD39+ TILs Significantly down-regulated genes in 4-1BBpos PD- Significantly up-regulated genes in 4-1BBpos PD-1high 1high CD39+ TILs CD39+ TILs (vs. 4-1BBneg PD-1high CD39+ TILs) (vs. 4-1BBneg PD-1high CD39+ TILs) 4-1BBneg cells 4-1BBpos cells 4-1BBneg cells 4-1BBpos cells Gene Pt 1 Pt 2 Pt 3 Pt 1 Pt 2 Pt 3 Gene Pt 1 Pt 2 Pt 3 Pt 1 Pt 2 Pt 3 name name FOSB 9.92496 9.80436 9.61180 9.76085 9.55460 8.47625 SPC25 2.82975 2.53201 1.83699 3.05751 3.79646 3.66172 5 9 4 4 3 9 8 5 5 6 7 PTGER4 7.26693 7.28598 7.19060 6.84482 6.61319 5.90088 GMNN 4.55416 5.05351 3.79176 4.95671 5.63164 4.93921 3 5 1 4 8 7 8 5 3 3 ABI3 6.24122 6.45779 6.27735 5.85419 5.81360 5.18958 ING3 4.78602 5.23034 4.49070 5.22964 5.62759 5.34212 1 3 5 6 6 2 3 6 5 8 7 1 LITAF 9.06689 9.17420 8.99478 8.79217 8.74965 8.08497 KIF14 4.58732 4.59859 4.62494 5.12658 5.70643 5.26241 4 4 9 8 7 5 7 9 3 7 4 ZNF10 4.90912 4.98940 4.74775 4.33282 4.40018 3.50431 CENPE 6.54529 6.55450 6.47696 6.84945 7.22848 6.96595 8 7 8 8 2 8 5 7 9 5 3 2 TNFAIP3 11.4201 11.2484 11.4068 11.1877 10.9900 10.5350 CDCA7 3.80074 4.20586 3.92787 4.43496 5.46931 4.40814 9 4 2 9 7 9 9 5 8 NAPA 5.94770 5.79864 5.64849