One-Way Analysis of Variance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

UCLA STAT 13 Comparison of Two Independent Samples
UCLA STAT 13 Introduction to Statistical Methods for the Life and Health Sciences Comparison of Two Instructor: Ivo Dinov, Independent Samples Asst. Prof. of Statistics and Neurology Teaching Assistants: Fred Phoa, Kirsten Johnson, Ming Zheng & Matilda Hsieh University of California, Los Angeles, Fall 2005 http://www.stat.ucla.edu/~dinov/courses_students.html Slide 1 Stat 13, UCLA, Ivo Dinov Slide 2 Stat 13, UCLA, Ivo Dinov Comparison of Two Independent Samples Comparison of Two Independent Samples z Many times in the sciences it is useful to compare two groups z Two Approaches for Comparison Male vs. Female Drug vs. Placebo Confidence Intervals NC vs. Disease we already know something about CI’s Hypothesis Testing Q: Different? this will be new µ µ z What seems like a reasonable way to 1 y 2 y2 σ 1 σ 1 s 2 compare two groups? 1 s2 z What parameter are we trying to estimate? Population 1 Sample 1 Population 2 Sample 2 Size n1 Size n2 Slide 3 Stat 13, UCLA, Ivo Dinov Slide 4 Stat 13, UCLA, Ivo Dinov − Comparison of Two Independent Samples Standard Error of y1 y2 y µ y − y µ − µ z RECALL: The sampling distribution of was centered at , z We know 1 2 estimates 1 2 and had a standard deviation of σ n z What we need to describe next is the precision of our estimate, SE()− y1 y2 − z We’ll start by describing the sampling distribution of y1 y2 Mean: µ1 – µ2 Standard deviation of σ 2 σ 2 s 2 s 2 1 + 2 = 1 + 2 = 2 + 2 SE()y − y SE1 SE2 n1 n2 1 2 n1 n2 z What seems like appropriate estimates for these quantities? Slide 5 Stat 13, UCLA, Ivo Dinov Slide 6 Stat 13, UCLA, Ivo Dinov 1 − − Standard Error of y1 y2 Standard Error of y1 y2 Example: A study is conducted to quantify the benefits of a new Example: Cholesterol medicine (cont’) cholesterol lowering medication. -
Statistical Analysis 8: Two-Way Analysis of Variance (ANOVA)
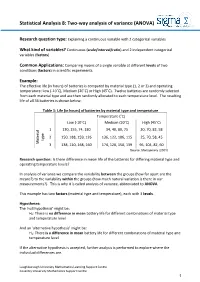
Statistical Analysis 8: Two-way analysis of variance (ANOVA) Research question type: Explaining a continuous variable with 2 categorical variables What kind of variables? Continuous (scale/interval/ratio) and 2 independent categorical variables (factors) Common Applications: Comparing means of a single variable at different levels of two conditions (factors) in scientific experiments. Example: The effective life (in hours) of batteries is compared by material type (1, 2 or 3) and operating temperature: Low (-10˚C), Medium (20˚C) or High (45˚C). Twelve batteries are randomly selected from each material type and are then randomly allocated to each temperature level. The resulting life of all 36 batteries is shown below: Table 1: Life (in hours) of batteries by material type and temperature Temperature (˚C) Low (-10˚C) Medium (20˚C) High (45˚C) 1 130, 155, 74, 180 34, 40, 80, 75 20, 70, 82, 58 2 150, 188, 159, 126 136, 122, 106, 115 25, 70, 58, 45 type Material 3 138, 110, 168, 160 174, 120, 150, 139 96, 104, 82, 60 Source: Montgomery (2001) Research question: Is there difference in mean life of the batteries for differing material type and operating temperature levels? In analysis of variance we compare the variability between the groups (how far apart are the means?) to the variability within the groups (how much natural variation is there in our measurements?). This is why it is called analysis of variance, abbreviated to ANOVA. This example has two factors (material type and temperature), each with 3 levels. Hypotheses: The 'null hypothesis' might be: H0: There is no difference in mean battery life for different combinations of material type and temperature level And an 'alternative hypothesis' might be: H1: There is a difference in mean battery life for different combinations of material type and temperature level If the alternative hypothesis is accepted, further analysis is performed to explore where the individual differences are. -

Sigmaplot 11: Now with Total Sigmastat Integration
SigmaPlot 11: Now with Total SigmaStat Integration Imagine my joy as I discovered a complete package of publication-quality graphics software with analytic and presentation tools John A. Wass, Ph.D., in: Scientific Computing International, Jan/Feb 2009 The SYSTAT people who market this product have thrown me a curve. For years, I have bemoaned the fact that most of the upgrade and development efforts that went into the SigmaPlot/SigmaStat software seemed to be biased to the plot side. When I observed that the new package was merely named SigmaPlot, and I further failed to find SigmaStat integration features (the stuff that connects the two programs), the Figure 1: SigmaPlot graphics and wizards, including the Quick Start natural conclusion seemed to be that the statistical Menu (upper right) and the graph program was jettisoned in favor of the graphics. wizard (bottom center) The above introductory narrative is intended to alert the reader to this editor’s long- time love affair with SigmaStat. It was the first statistical software that I used, (seemingly) the first to make a seamless transition from DOS to Windows, and the very first to offer that wonderful Wizard to we befuddled amateur statisticians. My introduction to SigmaPlot came much later, and use of that was only stimulated when the two became integrated. Later on, a pharmacology menu was added and the usage of the plotting software was greatly extended. Of course, the new version has added further graphics and helps to make an already useful program even easier to use. It is now a complete package of publication-quality graphics software with analytic and presentation tools. -

5. Dummy-Variable Regression and Analysis of Variance
Sociology 740 John Fox Lecture Notes 5. Dummy-Variable Regression and Analysis of Variance Copyright © 2014 by John Fox Dummy-Variable Regression and Analysis of Variance 1 1. Introduction I One of the limitations of multiple-regression analysis is that it accommo- dates only quantitative explanatory variables. I Dummy-variable regressors can be used to incorporate qualitative explanatory variables into a linear model, substantially expanding the range of application of regression analysis. c 2014 by John Fox Sociology 740 ° Dummy-Variable Regression and Analysis of Variance 2 2. Goals: I To show how dummy regessors can be used to represent the categories of a qualitative explanatory variable in a regression model. I To introduce the concept of interaction between explanatory variables, and to show how interactions can be incorporated into a regression model by forming interaction regressors. I To introduce the principle of marginality, which serves as a guide to constructing and testing terms in complex linear models. I To show how incremental I -testsareemployedtotesttermsindummy regression models. I To show how analysis-of-variance models can be handled using dummy variables. c 2014 by John Fox Sociology 740 ° Dummy-Variable Regression and Analysis of Variance 3 3. A Dichotomous Explanatory Variable I The simplest case: one dichotomous and one quantitative explanatory variable. I Assumptions: Relationships are additive — the partial effect of each explanatory • variable is the same regardless of the specific value at which the other explanatory variable is held constant. The other assumptions of the regression model hold. • I The motivation for including a qualitative explanatory variable is the same as for including an additional quantitative explanatory variable: to account more fully for the response variable, by making the errors • smaller; and to avoid a biased assessment of the impact of an explanatory variable, • as a consequence of omitting another explanatory variables that is relatedtoit. -

Analysis of Variance and Analysis of Variance and Design of Experiments of Experiments-I
Analysis of Variance and Design of Experimentseriments--II MODULE ––IVIV LECTURE - 19 EXPERIMENTAL DESIGNS AND THEIR ANALYSIS Dr. Shalabh Department of Mathematics and Statistics Indian Institute of Technology Kanpur 2 Design of experiment means how to design an experiment in the sense that how the observations or measurements should be obtained to answer a qqyuery inavalid, efficient and economical way. The desigggning of experiment and the analysis of obtained data are inseparable. If the experiment is designed properly keeping in mind the question, then the data generated is valid and proper analysis of data provides the valid statistical inferences. If the experiment is not well designed, the validity of the statistical inferences is questionable and may be invalid. It is important to understand first the basic terminologies used in the experimental design. Experimental unit For conducting an experiment, the experimental material is divided into smaller parts and each part is referred to as experimental unit. The experimental unit is randomly assigned to a treatment. The phrase “randomly assigned” is very important in this definition. Experiment A way of getting an answer to a question which the experimenter wants to know. Treatment Different objects or procedures which are to be compared in an experiment are called treatments. Sampling unit The object that is measured in an experiment is called the sampling unit. This may be different from the experimental unit. 3 Factor A factor is a variable defining a categorization. A factor can be fixed or random in nature. • A factor is termed as fixed factor if all the levels of interest are included in the experiment. -

Statistical Significance Testing in Information Retrieval:An Empirical
Statistical Significance Testing in Information Retrieval: An Empirical Analysis of Type I, Type II and Type III Errors Julián Urbano Harlley Lima Alan Hanjalic Delft University of Technology Delft University of Technology Delft University of Technology The Netherlands The Netherlands The Netherlands [email protected] [email protected] [email protected] ABSTRACT 1 INTRODUCTION Statistical significance testing is widely accepted as a means to In the traditional test collection based evaluation of Information assess how well a difference in effectiveness reflects an actual differ- Retrieval (IR) systems, statistical significance tests are the most ence between systems, as opposed to random noise because of the popular tool to assess how much noise there is in a set of evaluation selection of topics. According to recent surveys on SIGIR, CIKM, results. Random noise in our experiments comes from sampling ECIR and TOIS papers, the t-test is the most popular choice among various sources like document sets [18, 24, 30] or assessors [1, 2, 41], IR researchers. However, previous work has suggested computer but mainly because of topics [6, 28, 36, 38, 43]. Given two systems intensive tests like the bootstrap or the permutation test, based evaluated on the same collection, the question that naturally arises mainly on theoretical arguments. On empirical grounds, others is “how well does the observed difference reflect the real difference have suggested non-parametric alternatives such as the Wilcoxon between the systems and not just noise due to sampling of topics”? test. Indeed, the question of which tests we should use has accom- Our field can only advance if the published retrieval methods truly panied IR and related fields for decades now. -

THE ONE-SAMPLE Z TEST
10 THE ONE-SAMPLE z TEST Only the Lonely Difficulty Scale ☺ ☺ ☺ (not too hard—this is the first chapter of this kind, but youdistribute know more than enough to master it) or WHAT YOU WILL LEARN IN THIS CHAPTERpost, • Deciding when the z test for one sample is appropriate to use • Computing the observed z value • Interpreting the z value • Understandingcopy, what the z value means • Understanding what effect size is and how to interpret it not INTRODUCTION TO THE Do ONE-SAMPLE z TEST Lack of sleep can cause all kinds of problems, from grouchiness to fatigue and, in rare cases, even death. So, you can imagine health care professionals’ interest in seeing that their patients get enough sleep. This is especially the case for patients 186 Copyright ©2020 by SAGE Publications, Inc. This work may not be reproduced or distributed in any form or by any means without express written permission of the publisher. Chapter 10 ■ The One-Sample z Test 187 who are ill and have a real need for the healing and rejuvenating qualities that sleep brings. Dr. Joseph Cappelleri and his colleagues looked at the sleep difficul- ties of patients with a particular illness, fibromyalgia, to evaluate the usefulness of the Medical Outcomes Study (MOS) Sleep Scale as a measure of sleep problems. Although other analyses were completed, including one that compared a treat- ment group and a control group with one another, the important analysis (for our discussion) was the comparison of participants’ MOS scores with national MOS norms. Such a comparison between a sample’s mean score (the MOS score for par- ticipants in this study) and a population’s mean score (the norms) necessitates the use of a one-sample z test. -

Basic ES Computations, P. 1 BASIC EFFECT SIZE GUIDE with SPSS
Basic ES Computations, p. 1 BASIC EFFECT SIZE GUIDE WITH SPSS AND SAS SYNTAX Gregory J. Meyer, Robert E. McGrath, and Robert Rosenthal Last updated January 13, 2003 Pending: 1. Formulas for repeated measures/paired samples. (d = r / sqrt(1-r^2) 2. Explanation of 'set aside' lambda weights of 0 when computing focused contrasts. 3. Applications to multifactor designs. SECTION I: COMPUTING EFFECT SIZES FROM RAW DATA. I-A. The Pearson Correlation as the Effect Size I-A-1: Calculating Pearson's r From a Design With a Dimensional Variable and a Dichotomous Variable (i.e., a t-Test Design). I-A-2: Calculating Pearson's r From a Design With Two Dichotomous Variables (i.e., a 2 x 2 Chi-Square Design). I-A-3: Calculating Pearson's r From a Design With a Dimensional Variable and an Ordered, Multi-Category Variable (i.e., a Oneway ANOVA Design). I-A-4: Calculating Pearson's r From a Design With One Variable That Has 3 or More Ordered Categories and One Variable That Has 2 or More Ordered Categories (i.e., an Omnibus Chi-Square Design with df > 1). I-B. Cohen's d as the Effect Size I-B-1: Calculating Cohen's d From a Design With a Dimensional Variable and a Dichotomous Variable (i.e., a t-Test Design). SECTION II: COMPUTING EFFECT SIZES FROM THE OUTPUT OF STATISTICAL TESTS AND TRANSLATING ONE EFFECT SIZE TO ANOTHER. II-A. The Pearson Correlation as the Effect Size II-A-1: Pearson's r From t-Test Output Comparing Means Across Two Groups. -

Tests of Hypotheses Using Statistics
Tests of Hypotheses Using Statistics Adam Massey¤and Steven J. Millery Mathematics Department Brown University Providence, RI 02912 Abstract We present the various methods of hypothesis testing that one typically encounters in a mathematical statistics course. The focus will be on conditions for using each test, the hypothesis tested by each test, and the appropriate (and inappropriate) ways of using each test. We conclude by summarizing the di®erent tests (what conditions must be met to use them, what the test statistic is, and what the critical region is). Contents 1 Types of Hypotheses and Test Statistics 2 1.1 Introduction . 2 1.2 Types of Hypotheses . 3 1.3 Types of Statistics . 3 2 z-Tests and t-Tests 5 2.1 Testing Means I: Large Sample Size or Known Variance . 5 2.2 Testing Means II: Small Sample Size and Unknown Variance . 9 3 Testing the Variance 12 4 Testing Proportions 13 4.1 Testing Proportions I: One Proportion . 13 4.2 Testing Proportions II: K Proportions . 15 4.3 Testing r £ c Contingency Tables . 17 4.4 Incomplete r £ c Contingency Tables Tables . 18 5 Normal Regression Analysis 19 6 Non-parametric Tests 21 6.1 Tests of Signs . 21 6.2 Tests of Ranked Signs . 22 6.3 Tests Based on Runs . 23 ¤E-mail: [email protected] yE-mail: [email protected] 1 7 Summary 26 7.1 z-tests . 26 7.2 t-tests . 27 7.3 Tests comparing means . 27 7.4 Variance Test . 28 7.5 Proportions . 28 7.6 Contingency Tables . -

Statistical Significance
Statistical significance In statistical hypothesis testing,[1][2] statistical signif- 1.1 Related concepts icance (or a statistically significant result) is at- tained whenever the observed p-value of a test statis- The significance level α is the threshhold for p below tic is less than the significance level defined for the which the experimenter assumes the null hypothesis is study.[3][4][5][6][7][8][9] The p-value is the probability of false, and something else is going on. This means α is obtaining results at least as extreme as those observed, also the probability of mistakenly rejecting the null hy- given that the null hypothesis is true. The significance pothesis, if the null hypothesis is true.[22] level, α, is the probability of rejecting the null hypothe- Sometimes researchers talk about the confidence level γ sis, given that it is true.[10] This statistical technique for = (1 − α) instead. This is the probability of not rejecting testing the significance of results was developed in the the null hypothesis given that it is true. [23][24] Confidence early 20th century. levels and confidence intervals were introduced by Ney- In any experiment or observation that involves drawing man in 1937.[25] a sample from a population, there is always the possibil- ity that an observed effect would have occurred due to sampling error alone.[11][12] But if the p-value of an ob- 2 Role in statistical hypothesis test- served effect is less than the significance level, an inves- tigator may conclude that that effect reflects the charac- ing teristics of the -

Understanding Statistical Hypothesis Testing: the Logic of Statistical Inference
Review Understanding Statistical Hypothesis Testing: The Logic of Statistical Inference Frank Emmert-Streib 1,2,* and Matthias Dehmer 3,4,5 1 Predictive Society and Data Analytics Lab, Faculty of Information Technology and Communication Sciences, Tampere University, 33100 Tampere, Finland 2 Institute of Biosciences and Medical Technology, Tampere University, 33520 Tampere, Finland 3 Institute for Intelligent Production, Faculty for Management, University of Applied Sciences Upper Austria, Steyr Campus, 4040 Steyr, Austria 4 Department of Mechatronics and Biomedical Computer Science, University for Health Sciences, Medical Informatics and Technology (UMIT), 6060 Hall, Tyrol, Austria 5 College of Computer and Control Engineering, Nankai University, Tianjin 300000, China * Correspondence: [email protected]; Tel.: +358-50-301-5353 Received: 27 July 2019; Accepted: 9 August 2019; Published: 12 August 2019 Abstract: Statistical hypothesis testing is among the most misunderstood quantitative analysis methods from data science. Despite its seeming simplicity, it has complex interdependencies between its procedural components. In this paper, we discuss the underlying logic behind statistical hypothesis testing, the formal meaning of its components and their connections. Our presentation is applicable to all statistical hypothesis tests as generic backbone and, hence, useful across all application domains in data science and artificial intelligence. Keywords: hypothesis testing; machine learning; statistics; data science; statistical inference 1. Introduction We are living in an era that is characterized by the availability of big data. In order to emphasize the importance of this, data have been called the ‘oil of the 21st Century’ [1]. However, for dealing with the challenges posed by such data, advanced analysis methods are needed. -

ANALYSIS of VARIANCE and MISSING OBSERVATIONS in COMPLETELY RANDOMIZED, RANDOMIZED BLOCKS and LATIN SQUARE DESIGNS a Thesis
ANALYSIS OF VARIANCE AND MISSING OBSERVATIONS IN COMPLETELY RANDOMIZED, RANDOMIZED BLOCKS AND LATIN SQUARE DESIGNS A Thesis Presented to The Department of Mathematics Kansas State Teachers College, Emporia, Kansas In Partial Fulfillment of the Requirements for the Degree Master of Science by Kiritkumar K. Talati May 1972 c; , ACKNOWLEDGEMENTS Sincere thanks and gratitude is expressed to Dr. John Burger for his assistance, patience, and his prompt attention in all directions in preparing this paper. A special note of appreciation goes to Dr. Marion Emerson, Dr. Thomas Davis, Dr. George Poole, Dr. Darrell Wood, who rendered assistance in the research of this paper. TABLE OF CONTENTS CHAPTER I. INTRODUCTION • • • • • • • • • • • • • • • • 1 A. Preliminary Consideration. • • • • • • • 1 B. Purpose and Assumptions of the Analysis of Variance • • • • • • • • •• 1 C. Analysis of Covariance • • • • • • • • • 2 D. Definitions. • • • • • • • • • • • • • • ) E. Organization of Paper. • • • • • • • • • 4 II. COMPLETELY RANDOMIZED DESIGN • • • • • • • • 5 A. Description............... 5 B. Randomization. • • • • • • • • • • • • • 5 C. Problem and Computations •••••••• 6 D. Conclusion and Further Applications. •• 10 III. RANDOMIZED BLOCK DESIGN. • • • • • • • • • • 12 A. Description............... 12 B. Randomization. • • • • • • • • • • • • • 1) C. Problem and Statistical Analysis • • • • 1) D. Efficiency of Randomized Block Design as Compared to Completely Randomized Design. • • • • • • • • • •• 20 E. Missing Observations • • • • • • • • •• 21 F.