Molecular Systematics of the Phoxinin Genus Pteronotropis (Otophysi: Cypriniformes)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BLUENOSE SHINER Scientific Name: Pteronotropis Welaka Evermann
Common Name: BLUENOSE SHINER Scientific Name: Pteronotropis welaka Evermann and Kendall Other Commonly Used Names: none Previously Used Scientific Names: Notropis welaka Family: Cyprinidae Rarity Ranks: G3G4/S1 State Legal Status: Threatened Federal Legal Status: Not Listed Description: The bluenose shiner is a slender minnow with a compressed body, a pointed snout and a terminal to subterminal mouth. Adults reach 53 mm (2.1 in) standard length. The first ray of the dorsal fin (i.e., the origin) is distinctly posterior to the first ray of the pelvic fin. There are 3-13 pored scales on the lateral line, 8-9 anal fin rays, and a typical pharyngeal tooth count formula of 1-4-4-1 (occasionally 0-4-4-0). A wide, dark lateral stripe runs along the length of the body between the snout and the caudal spot and is often flecked with silvery scales in both males and females. A thin, light-yellow stripe runs just above the black lateral stripe. The oblong caudal spot extends onto the median rays of the caudal fin and is bordered above and below by de-pigmented areas. The dorsum is dusky olive-brown and the venter is pale. This species exhibits striking sexual dimorphism, which includes a bright blue snout and greatly enlarged dorsal, pelvic, and anal fins on breeding males. The dorsal fin becomes black with a pale band near its base and the anal and pelvic fins turn white to golden yellow with contrasting black pigment within the middle of each fin. These characteristics develop gradually with age and many males will exhibit only partial development of nuptial male morphology and color patterns (e.g., a blue snout, but not greatly enlarged fins). -

Environmental Sensitivity Index Guidelines Version 2.0
NOAA Technical Memorandum NOS ORCA 115 Environmental Sensitivity Index Guidelines Version 2.0 October 1997 Seattle, Washington noaa NATIONAL OCEANIC AND ATMOSPHERIC ADMINISTRATION National Ocean Service Office of Ocean Resources Conservation and Assessment National Ocean Service National Oceanic and Atmospheric Administration U.S. Department of Commerce The Office of Ocean Resources Conservation and Assessment (ORCA) provides decisionmakers comprehensive, scientific information on characteristics of the oceans, coastal areas, and estuaries of the United States of America. The information ranges from strategic, national assessments of coastal and estuarine environmental quality to real-time information for navigation or hazardous materials spill response. Through its National Status and Trends (NS&T) Program, ORCA uses uniform techniques to monitor toxic chemical contamination of bottom-feeding fish, mussels and oysters, and sediments at about 300 locations throughout the United States. A related NS&T Program of directed research examines the relationships between contaminant exposure and indicators of biological responses in fish and shellfish. Through the Hazardous Materials Response and Assessment Division (HAZMAT) Scientific Support Coordination program, ORCA provides critical scientific support for planning and responding to spills of oil or hazardous materials into coastal environments. Technical guidance includes spill trajectory predictions, chemical hazard analyses, and assessments of the sensitivity of marine and estuarine environments to spills. To fulfill the responsibilities of the Secretary of Commerce as a trustee for living marine resources, HAZMAT’s Coastal Resource Coordination program provides technical support to the U.S. Environmental Protection Agency during all phases of the remedial process to protect the environment and restore natural resources at hundreds of waste sites each year. -

Endangered Species
FEATURE: ENDANGERED SPECIES Conservation Status of Imperiled North American Freshwater and Diadromous Fishes ABSTRACT: This is the third compilation of imperiled (i.e., endangered, threatened, vulnerable) plus extinct freshwater and diadromous fishes of North America prepared by the American Fisheries Society’s Endangered Species Committee. Since the last revision in 1989, imperilment of inland fishes has increased substantially. This list includes 700 extant taxa representing 133 genera and 36 families, a 92% increase over the 364 listed in 1989. The increase reflects the addition of distinct populations, previously non-imperiled fishes, and recently described or discovered taxa. Approximately 39% of described fish species of the continent are imperiled. There are 230 vulnerable, 190 threatened, and 280 endangered extant taxa, and 61 taxa presumed extinct or extirpated from nature. Of those that were imperiled in 1989, most (89%) are the same or worse in conservation status; only 6% have improved in status, and 5% were delisted for various reasons. Habitat degradation and nonindigenous species are the main threats to at-risk fishes, many of which are restricted to small ranges. Documenting the diversity and status of rare fishes is a critical step in identifying and implementing appropriate actions necessary for their protection and management. Howard L. Jelks, Frank McCormick, Stephen J. Walsh, Joseph S. Nelson, Noel M. Burkhead, Steven P. Platania, Salvador Contreras-Balderas, Brady A. Porter, Edmundo Díaz-Pardo, Claude B. Renaud, Dean A. Hendrickson, Juan Jacobo Schmitter-Soto, John Lyons, Eric B. Taylor, and Nicholas E. Mandrak, Melvin L. Warren, Jr. Jelks, Walsh, and Burkhead are research McCormick is a biologist with the biologists with the U.S. -

Aquatic Fish Report
Aquatic Fish Report Acipenser fulvescens Lake St urgeon Class: Actinopterygii Order: Acipenseriformes Family: Acipenseridae Priority Score: 27 out of 100 Population Trend: Unknown Gobal Rank: G3G4 — Vulnerable (uncertain rank) State Rank: S2 — Imperiled in Arkansas Distribution Occurrence Records Ecoregions where the species occurs: Ozark Highlands Boston Mountains Ouachita Mountains Arkansas Valley South Central Plains Mississippi Alluvial Plain Mississippi Valley Loess Plains Acipenser fulvescens Lake Sturgeon 362 Aquatic Fish Report Ecobasins Mississippi River Alluvial Plain - Arkansas River Mississippi River Alluvial Plain - St. Francis River Mississippi River Alluvial Plain - White River Mississippi River Alluvial Plain (Lake Chicot) - Mississippi River Habitats Weight Natural Littoral: - Large Suitable Natural Pool: - Medium - Large Optimal Natural Shoal: - Medium - Large Obligate Problems Faced Threat: Biological alteration Source: Commercial harvest Threat: Biological alteration Source: Exotic species Threat: Biological alteration Source: Incidental take Threat: Habitat destruction Source: Channel alteration Threat: Hydrological alteration Source: Dam Data Gaps/Research Needs Continue to track incidental catches. Conservation Actions Importance Category Restore fish passage in dammed rivers. High Habitat Restoration/Improvement Restrict commercial harvest (Mississippi River High Population Management closed to harvest). Monitoring Strategies Monitor population distribution and abundance in large river faunal surveys in cooperation -
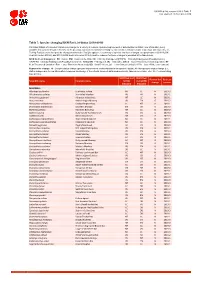
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Molecular Systematics of Western North American Cyprinids (Cypriniformes: Cyprinidae)
Zootaxa 3586: 281–303 (2012) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2012 · Magnolia Press Article ISSN 1175-5334 (online edition) urn:lsid:zoobank.org:pub:0EFA9728-D4BB-467E-A0E0-0DA89E7E30AD Molecular systematics of western North American cyprinids (Cypriniformes: Cyprinidae) SUSANA SCHÖNHUTH 1, DENNIS K. SHIOZAWA 2, THOMAS E. DOWLING 3 & RICHARD L. MAYDEN 1 1 Department of Biology, Saint Louis University, 3507 Laclede Avenue, St. Louis, MO 63103, USA. E-mail S.S: [email protected] ; E-mail RLM: [email protected] 2 Department of Biology and Curator of Fishes, Monte L. Bean Life Science Museum, Brigham Young University, Provo, UT 84602, USA. E-mail: [email protected] 3 School of Life Sciences, Arizona State University, Tempe, AZ 85287-4501, USA. E-mail: [email protected] Abstract The phylogenetic or evolutionary relationships of species of Cypriniformes, as well as their classification, is in a era of flux. For the first time ever, the Order, and constituent Families are being examined for relationships within a phylogenetic context. Relevant findings as to sister-group relationships are largely being inferred from analyses of both mitochondrial and nuclear DNA sequences. Like the vast majority of Cypriniformes, due to an overall lack of any phylogenetic investigation of these fishes since Hennig’s transformation of the discipline, changes in hypotheses of relationships and a natural classification of the species should not be of surprise to anyone. Basically, for most taxa no properly supported phylogenetic hypothesis has ever been done; and this includes relationships with reasonable taxon and character sampling of even families and subfamilies. -

Biobasics Contents
Illinois Biodiversity Basics a biodiversity education program of Illinois Department of Natural Resources Chicago Wilderness World Wildlife Fund Adapted from Biodiversity Basics, © 1999, a publication of World Wildlife Fund’s Windows on the Wild biodiversity education program. For more information see <www.worldwildlife.org/windows>. Table of Contents About Illinois Biodiversity Basics ................................................................................................................. 2 Biodiversity Background ............................................................................................................................... 4 Biodiversity of Illinois CD-ROM series ........................................................................................................ 6 Activities Section 1: What is Biodiversity? ...................................................................................................... 7 Activity 1-1: What’s Your Biodiversity IQ?.................................................................... 8 Activity 1-2: Sizing Up Species .................................................................................... 19 Activity 1-3: Backyard BioBlitz.................................................................................... 31 Activity 1-4: The Gene Scene ....................................................................................... 43 Section 2: Why is Biodiversity Important? .................................................................................... 61 Activity -

Abstracts Part 1
375 Poster Session I, Event Center – The Snowbird Center, Friday 26 July 2019 Maria Sabando1, Yannis Papastamatiou1, Guillaume Rieucau2, Darcy Bradley3, Jennifer Caselle3 1Florida International University, Miami, FL, USA, 2Louisiana Universities Marine Consortium, Chauvin, LA, USA, 3University of California, Santa Barbara, Santa Barbara, CA, USA Reef Shark Behavioral Interactions are Habitat Specific Dominance hierarchies and competitive behaviors have been studied in several species of animals that includes mammals, birds, amphibians, and fish. Competition and distribution model predictions vary based on dominance hierarchies, but most assume differences in dominance are constant across habitats. More recent evidence suggests dominance and competitive advantages may vary based on habitat. We quantified dominance interactions between two species of sharks Carcharhinus amblyrhynchos and Carcharhinus melanopterus, across two different habitats, fore reef and back reef, at a remote Pacific atoll. We used Baited Remote Underwater Video (BRUV) to observe dominance behaviors and quantified the number of aggressive interactions or bites to the BRUVs from either species, both separately and in the presence of one another. Blacktip reef sharks were the most abundant species in either habitat, and there was significant negative correlation between their relative abundance, bites on BRUVs, and the number of grey reef sharks. Although this trend was found in both habitats, the decline in blacktip abundance with grey reef shark presence was far more pronounced in fore reef habitats. We show that the presence of one shark species may limit the feeding opportunities of another, but the extent of this relationship is habitat specific. Future competition models should consider habitat-specific dominance or competitive interactions. -

Checklist of the Inland Fishes of Louisiana
Southeastern Fishes Council Proceedings Volume 1 Number 61 2021 Article 3 March 2021 Checklist of the Inland Fishes of Louisiana Michael H. Doosey University of New Orelans, [email protected] Henry L. Bart Jr. Tulane University, [email protected] Kyle R. Piller Southeastern Louisiana Univeristy, [email protected] Follow this and additional works at: https://trace.tennessee.edu/sfcproceedings Part of the Aquaculture and Fisheries Commons, and the Biodiversity Commons Recommended Citation Doosey, Michael H.; Bart, Henry L. Jr.; and Piller, Kyle R. (2021) "Checklist of the Inland Fishes of Louisiana," Southeastern Fishes Council Proceedings: No. 61. Available at: https://trace.tennessee.edu/sfcproceedings/vol1/iss61/3 This Original Research Article is brought to you for free and open access by Volunteer, Open Access, Library Journals (VOL Journals), published in partnership with The University of Tennessee (UT) University Libraries. This article has been accepted for inclusion in Southeastern Fishes Council Proceedings by an authorized editor. For more information, please visit https://trace.tennessee.edu/sfcproceedings. Checklist of the Inland Fishes of Louisiana Abstract Since the publication of Freshwater Fishes of Louisiana (Douglas, 1974) and a revised checklist (Douglas and Jordan, 2002), much has changed regarding knowledge of inland fishes in the state. An updated reference on Louisiana’s inland and coastal fishes is long overdue. Inland waters of Louisiana are home to at least 224 species (165 primarily freshwater, 28 primarily marine, and 31 euryhaline or diadromous) in 45 families. This checklist is based on a compilation of fish collections records in Louisiana from 19 data providers in the Fishnet2 network (www.fishnet2.net). -

Notropis Cahabae)
Cahaba Shiner (Notropis cahabae) 5-Year Review: Summary and Evaluation Cahaba Shiner, from Fishes of Alabama and the Mobile Basin, Mettee, O’Neil, and Pierson 1987. U.S. Fish and Wildlife Service Southeast Region Mississippi Ecological Services Field Office Jackson, Mississippi 1 5-YEAR REVIEW Cahaba shiner (Notropis cahabae) I. GENERAL INFORMATION A. Methodology used to complete the review: In conducting this 5-year review, we relied on the best available information pertaining to historical and current distributions, life histories, and habitats of this species. We announced initiation of this review and requested information in a published Federal Register notice with a 60-day comment period (74 FR 31972). We conducted an internet search, reviewed all information in our files, and solicited information from knowledgeable individuals familiar with this species including those associated with academia and State conservation programs. Specific sources included the final rule listing this species under the Endangered Species Act; the Recovery Plan; peer reviewed scientific publications; unpublished field observations by the U.S. Fish and Wildlife Service, State and other experienced biologists; unpublished survey reports; and notes and communications from other qualified biologists or experts. The completed draft was sent to other associated Service offices and peer reviewers. Comments were evaluated and incorporated, as appropriate, into this final document (see Appendix A). No public comments were received. B. Reviewers Lead Region – Southeast Region: Kelly Bibb, 404-679-7132 Lead Field Office – Mississippi Ecological Services Field Office: Daniel J. Drennen, 601-321-1127 Cooperating Field Office – Alabama Ecological Services Field Office: Jeff Powell, 251-441-5858. C. -

Cypriniformes: Cyprinidae)
Zootaxa 3586: 281–303 (2012) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2012 · Magnolia Press Article ISSN 1175-5334 (online edition) urn:lsid:zoobank.org:pub:0EFA9728-D4BB-467E-A0E0-0DA89E7E30AD Molecular systematics of western North American cyprinids (Cypriniformes: Cyprinidae) SUSANA SCHÖNHUTH 1, DENNIS K. SHIOZAWA 2, THOMAS E. DOWLING 3 & RICHARD L. MAYDEN 1 1 Department of Biology, Saint Louis University, 3507 Laclede Avenue, St. Louis, MO 63103, USA. E-mail S.S: [email protected] ; E-mail RLM: [email protected] 2 Department of Biology and Curator of Fishes, Monte L. Bean Life Science Museum, Brigham Young University, Provo, UT 84602, USA. E-mail: [email protected] 3 School of Life Sciences, Arizona State University, Tempe, AZ 85287-4501, USA. E-mail: [email protected] Abstract The phylogenetic or evolutionary relationships of species of Cypriniformes, as well as their classification, is in a era of flux. For the first time ever, the Order, and constituent Families are being examined for relationships within a phylogenetic context. Relevant findings as to sister-group relationships are largely being inferred from analyses of both mitochondrial and nuclear DNA sequences. Like the vast majority of Cypriniformes, due to an overall lack of any phylogenetic investigation of these fishes since Hennig’s transformation of the discipline, changes in hypotheses of relationships and a natural classification of the species should not be of surprise to anyone. Basically, for most taxa no properly supported phylogenetic hypothesis has ever been done; and this includes relationships with reasonable taxon and character sampling of even families and subfamilies. -

Molecular Systematics of the Phoxinin Genus Pteronotropis (Otophysi: Cypriniformes)
Hindawi Publishing Corporation BioMed Research International Volume 2015, Article ID 298658, 8 pages http://dx.doi.org/10.1155/2015/298658 Research Article Molecular Systematics of the Phoxinin Genus Pteronotropis (Otophysi: Cypriniformes) Richard L. Mayden1 and Jason S. Allen2 1 Department of Biology, Saint Louis University, 3507 Laclede Avenue, St. Louis, MO 63103, USA 2Department of Biology, Saint Louis Community College, Meramec Campus, 11333 Big Bend Road, St. Louis, MO 63122, USA Correspondence should be addressed to Richard L. Mayden; [email protected] Received 22 June 2014; Revised 13 December 2014; Accepted 13 December 2014 Academic Editor: Vassily Lyubetsky Copyright © 2015 R. L. Mayden and J. S. Allen. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The genus Pteronotropis is widely distributed along the gulf slope of eastern North America from Louisiana to Florida and rivers in South Carolina along the Atlantic slope. Pteronotropis have very distinctive, flamboyant coloration. The habitats most frequently associated with these species include heavily vegetated backwater bayous to small sluggish or flowing tannin-stained streams. Although Pteronotropis is recognized as a valid genus, no phylogenetic analysis of all the species has corroborated its monophyly. In recent years, four additional species have been either described or elevated from synonymy: P. me rlini , P. grandipinnis, P. stone i ,and P. metal li c u s , with the wide-ranging P. hy ps el opte r u s complex. To examine relationships within this genus and test its monophyly, phylogenetic analyses were conducted using two nuclear genes, recombination activating gene 1, RAG1, and the first intron of S7 ribosomal protein gene in both maximum parsimony and Bayesian analyses.