New Open Source Software for Building Molecular Dynamics Systems
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Olaseni Sode
Olaseni Sode Work Address: 5735 S. Ellis Ave. [email protected] Searle 231 +1 (314) 856-2373 The University of Chicago Chicago, IL 60615 Home Address: 1119 E Hyde Park Blvd [email protected] Apt. 3E +1 (314) 856-2373 Chicago, IL 60615 http://www.olaseniosode.com Education University of Illinois at Urbana-Champaign Urbana, IL Ph.D. in Chemistry September 2012 – Supervisor: Professor So Hirata – Thesis title: A Theoretical Study of Molecular Crystals – Transferred from the University of Florida in 2010 with Prof. Hirata Morehouse College Atlanta, GA B.S. & B.A. in Chemistry and French December 2006 – Studied at the Université de Nantes, Nantes, France (2004-2005) Research Experience The University of Chicago Chicago, IL Postdoctoral Research – research advisor: Dr. Gregory A. Voth 2012–present – Studied proposed proton transport minimum free-energy pathways in [FeFe]-hydrogenase via molecular dynamics simulations and the multistate empirical valence bond technique. – Collaborated to develop the multiscale string method to accelerate refinement of chemical reaction pathways of adenosine triphosphate (ATP) hydrolysis in globular and filamentous actin proteins. University of California, San Diego La Jolla, CA Visiting Scholar – research advisor: Dr. Francesco Paesani 2014, 2015 – Developed and implemented a reactive molecular dynamics scheme for condensed phase systems of the hydrated hydronium ions. University of Illinois at Urbana-Champaign Urbana, IL Doctoral Research – research advisor: Dr. So Hirata 2010-2012 – Characterized the vibrational structure of extended systems including infrared and Raman-active vibrations, as well as phonon dispersions and densities of states in the first Brillouin zone. – Developed zero-point vibrational energy analysis scheme for molecular clusters using a fragmented, local basis scheme. -

Pysimm: a Python Package for Simulation of Molecular Systems
SoftwareX 6 (2016) 7–12 Contents lists available at ScienceDirect SoftwareX journal homepage: www.elsevier.com/locate/softx pysimm: A python package for simulation of molecular systems Michael E. Fortunato a, Coray M. Colina a,b,∗ a Department of Chemistry, University of Florida, Gainesville, FL 32611, United States b Department of Materials Science and Engineering and Nuclear Engineering, University of Florida, Gainesville, FL 32611, United States article info a b s t r a c t Article history: In this work, we present pysimm, a python package designed to facilitate structure generation, simulation, Received 15 August 2016 and modification of molecular systems. pysimm provides a collection of simulation tools and smooth Received in revised form integration with highly optimized third party software. Abstraction layers enable a standardized 1 December 2016 methodology to assign various force field models to molecular systems and perform simple simulations. Accepted 5 December 2016 These features have allowed pysimm to aid the rapid development of new applications specifically in the area of amorphous polymer simulations. Keywords: ' 2016 The Authors. Published by Elsevier B.V. Amorphous polymers Molecular simulation This is an open access article under the CC BY license Python (http://creativecommons.org/licenses/by/4.0/). Code metadata Current code version v0.1 Permanent link to code/repository used for this code version https://github.com/ElsevierSoftwareX/SOFTX-D-16-00070 Legal Code License MIT Code versioning system used git Software code languages, tools, and services used python2.7 Compilation requirements, operating environments & Linux dependencies If available Link to developer documentation/manual http://pysimm.org/documentation/ Support email for questions [email protected] 1. -

Open Babel Documentation Release 2.3.1
Open Babel Documentation Release 2.3.1 Geoffrey R Hutchison Chris Morley Craig James Chris Swain Hans De Winter Tim Vandermeersch Noel M O’Boyle (Ed.) December 05, 2011 Contents 1 Introduction 3 1.1 Goals of the Open Babel project ..................................... 3 1.2 Frequently Asked Questions ....................................... 4 1.3 Thanks .................................................. 7 2 Install Open Babel 9 2.1 Install a binary package ......................................... 9 2.2 Compiling Open Babel .......................................... 9 3 obabel and babel - Convert, Filter and Manipulate Chemical Data 17 3.1 Synopsis ................................................. 17 3.2 Options .................................................. 17 3.3 Examples ................................................. 19 3.4 Differences between babel and obabel .................................. 21 3.5 Format Options .............................................. 22 3.6 Append property values to the title .................................... 22 3.7 Filtering molecules from a multimolecule file .............................. 22 3.8 Substructure and similarity searching .................................. 25 3.9 Sorting molecules ............................................ 25 3.10 Remove duplicate molecules ....................................... 25 3.11 Aliases for chemical groups ....................................... 26 4 The Open Babel GUI 29 4.1 Basic operation .............................................. 29 4.2 Options ................................................. -

Open Data, Open Source, and Open Standards in Chemistry: the Blue Obelisk Five Years On" Journal of Cheminformatics Vol
Oral Roberts University Digital Showcase College of Science and Engineering Faculty College of Science and Engineering Research and Scholarship 10-14-2011 Open Data, Open Source, and Open Standards in Chemistry: The lueB Obelisk five years on Andrew Lang Noel M. O'Boyle Rajarshi Guha National Institutes of Health Egon Willighagen Maastricht University Samuel Adams See next page for additional authors Follow this and additional works at: http://digitalshowcase.oru.edu/cose_pub Part of the Chemistry Commons Recommended Citation Andrew Lang, Noel M O'Boyle, Rajarshi Guha, Egon Willighagen, et al.. "Open Data, Open Source, and Open Standards in Chemistry: The Blue Obelisk five years on" Journal of Cheminformatics Vol. 3 Iss. 37 (2011) Available at: http://works.bepress.com/andrew-sid-lang/ 19/ This Article is brought to you for free and open access by the College of Science and Engineering at Digital Showcase. It has been accepted for inclusion in College of Science and Engineering Faculty Research and Scholarship by an authorized administrator of Digital Showcase. For more information, please contact [email protected]. Authors Andrew Lang, Noel M. O'Boyle, Rajarshi Guha, Egon Willighagen, Samuel Adams, Jonathan Alvarsson, Jean- Claude Bradley, Igor Filippov, Robert M. Hanson, Marcus D. Hanwell, Geoffrey R. Hutchison, Craig A. James, Nina Jeliazkova, Karol M. Langner, David C. Lonie, Daniel M. Lowe, Jerome Pansanel, Dmitry Pavlov, Ola Spjuth, Christoph Steinbeck, Adam L. Tenderholt, Kevin J. Theisen, and Peter Murray-Rust This article is available at Digital Showcase: http://digitalshowcase.oru.edu/cose_pub/34 Oral Roberts University From the SelectedWorks of Andrew Lang October 14, 2011 Open Data, Open Source, and Open Standards in Chemistry: The Blue Obelisk five years on Andrew Lang Noel M O'Boyle Rajarshi Guha, National Institutes of Health Egon Willighagen, Maastricht University Samuel Adams, et al. -

A Study on Cheminformatics and Its Applications on Modern Drug Discovery
Available online at www.sciencedirect.com Procedia Engineering 38 ( 2012 ) 1264 – 1275 Internatio na l Conference on Modeling Optimisatio n and Computing (ICMOC 2012) A Study on Cheminformatics and its Applications on Modern Drug Discovery B.Firdaus Begama and Dr. J.Satheesh Kumarb aResearch Scholar, Bharathiar University, Coimbatore, India, [email protected] bAssistant Professor, Bharathiar University, Coimbatore, India, [email protected] Abstract Discovering drugs to a disease is still a challenging task for medical researchers due to the complex structures of biomolecules which are responsible for disease such as AIDS, Cancer, Autism, Alzimear etc. Design and development of new efficient anti-drugs for the disease without any side effects are becoming mandatory in the recent history of human life cycle due to changes in various factors which includes food habit, environmental and migration in human life style. Cheminformaticds deals with discovering drugs based in modern drug discovery techniques which in turn rectifies complex issues in traditional drug discovery system. Cheminformatics tools, helps medical chemist for better understanding of complex structures of chemical compounds. Cheminformatics is a new emerging interdisciplinary field which primarily aims to discover Novel Chemical Entities [NCE] which ultimately results in design of new molecule [chemical data]. It also plays an important role for collecting, storing and analysing the chemical data. This paper focuses on cheminformatics and its applications on drug discovery and modern drug discovery techniques which helps chemist and medical researchers for finding solution to the complex disease. © 2012 Published by Elsevier Ltd. Selection and/or peer-review under responsibility of Noorul Islam Centre for Higher Education. -

Computer-Assisted Catalyst Development Via Automated Modelling of Conformationally Complex Molecules
www.nature.com/scientificreports OPEN Computer‑assisted catalyst development via automated modelling of conformationally complex molecules: application to diphosphinoamine ligands Sibo Lin1*, Jenna C. Fromer2, Yagnaseni Ghosh1, Brian Hanna1, Mohamed Elanany3 & Wei Xu4 Simulation of conformationally complicated molecules requires multiple levels of theory to obtain accurate thermodynamics, requiring signifcant researcher time to implement. We automate this workfow using all open‑source code (XTBDFT) and apply it toward a practical challenge: diphosphinoamine (PNP) ligands used for ethylene tetramerization catalysis may isomerize (with deleterious efects) to iminobisphosphines (PPNs), and a computational method to evaluate PNP ligand candidates would save signifcant experimental efort. We use XTBDFT to calculate the thermodynamic stability of a wide range of conformationally complex PNP ligands against isomeriation to PPN (ΔGPPN), and establish a strong correlation between ΔGPPN and catalyst performance. Finally, we apply our method to screen novel PNP candidates, saving signifcant time by ruling out candidates with non‑trivial synthetic routes and poor expected catalytic performance. Quantum mechanical methods with high energy accuracy, such as density functional theory (DFT), can opti- mize molecular input structures to a nearby local minimum, but calculating accurate reaction thermodynamics requires fnding global minimum energy structures1,2. For simple molecules, expert intuition can identify a few minima to focus study on, but an alternative approach must be considered for more complex molecules or to eventually fulfl the dream of autonomous catalyst design 3,4: the potential energy surface must be frst surveyed with a computationally efcient method; then minima from this survey must be refned using slower, more accurate methods; fnally, for molecules possessing low-frequency vibrational modes, those modes need to be treated appropriately to obtain accurate thermodynamic energies 5–7. -

Designing Universal Chemical Markup (UCM) Through the Reusable Methodology Based on Analyzing Existing Related Formats
Designing Universal Chemical Markup (UCM) through the reusable methodology based on analyzing existing related formats Background: In order to design concepts for a new general-purpose chemical format we analyzed the strengths and weaknesses of current formats for common chemical data. While the new format is discussed more in the next article, here we describe our software s t tools and two stage analysis procedure that supplied the necessary information for the n i r development. The chemical formats analyzed in both stages were: CDX, CDXML, CML, P CTfile and XDfile. In addition the following formats were included in the first stage only: e r P CIF, InChI, NCBI ASN.1, NCBI XML, PDB, PDBx/mmCIF, PDBML, SMILES, SLN and Mol2. Results: A two stage analysis process devised for both XML (Extensible Markup Language) and non-XML formats enabled us to verify if and how potential advantages of XML are utilized in the widely used general-purpose chemical formats. In the first stage we accumulated information about analyzed formats and selected the formats with the most general-purpose chemical functionality for the second stage. During the second stage our set of software quality requirements was used to assess the benefits and issues of selected formats. Additionally, the detailed analysis of XML formats structure in the second stage helped us to identify concepts in those formats. Using these concepts we came up with the concise structure for a new chemical format, which is designed to provide precise built-in validation capabilities and aims to avoid the potential issues of analyzed formats. -
Biovia Materials Studio Visualizer Datasheet
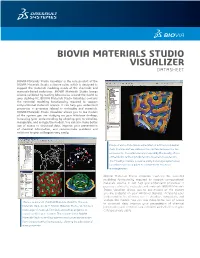
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

Open Data, Open Source and Open Standards in Chemistry: the Blue Obelisk five Years On
Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk ¯ve years on Noel M O'Boyle¤1 , Rajarshi Guha2 , Egon L Willighagen3 , Samuel E Adams4 , Jonathan Alvarsson5 , Richard L Apodaca6 , Jean-Claude Bradley7 , Igor V Filippov8 , Robert M Hanson9 , Marcus D Hanwell10 , Geo®rey R Hutchison11 , Craig A James12 , Nina Jeliazkova13 , Andrew SID Lang14 , Karol M Langner15 , David C Lonie16 , Daniel M Lowe4 , J¶er^omePansanel17 , Dmitry Pavlov18 , Ola Spjuth5 , Christoph Steinbeck19 , Adam L Tenderholt20 , Kevin J Theisen21 , Peter Murray-Rust4 1Analytical and Biological Chemistry Research Facility, Cavanagh Pharmacy Building, University College Cork, College Road, Cork, Co. Cork, Ireland 2NIH Center for Translational Therapeutics, 9800 Medical Center Drive, Rockville, MD 20878, USA 3Division of Molecular Toxicology, Institute of Environmental Medicine, Nobels vaeg 13, Karolinska Institutet, 171 77 Stockholm, Sweden 4Unilever Centre for Molecular Sciences Informatics, Department of Chemistry, University of Cambridge, Lens¯eld Road, CB2 1EW, UK 5Department of Pharmaceutical Biosciences, Uppsala University, Box 591, 751 24 Uppsala, Sweden 6Metamolecular, LLC, 8070 La Jolla Shores Drive #464, La Jolla, CA 92037, USA 7Department of Chemistry, Drexel University, 32nd and Chestnut streets, Philadelphia, PA 19104, USA 8Chemical Biology Laboratory, Basic Research Program, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, USA 9St. Olaf College, 1520 St. Olaf Ave., North¯eld, MN 55057, USA 10Kitware, Inc., 28 Corporate Drive, Clifton Park, NY 12065, USA 11Department of Chemistry, University of Pittsburgh, 219 Parkman Avenue, Pittsburgh, PA 15260, USA 12eMolecules Inc., 380 Stevens Ave., Solana Beach, California 92075, USA 13Ideaconsult Ltd., 4.A.Kanchev str., So¯a 1000, Bulgaria 14Department of Engineering, Computer Science, Physics, and Mathematics, Oral Roberts University, 7777 S. -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12 -

Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins
Article Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins Marcus Purse 1, Grace Edmund 1, Stephen Hall 1, Brendan Howlin 1,* , Ian Hamerton 2 and Stephen Till 3 1 Department of Chemistry, Faculty of Engineering and Physical Sciences, University of Surrey, Guilford, Surrey GU2 7XH, UK; [email protected] (M.P.); [email protected] (G.E.); [email protected] (S.H.) 2 Bristol Composites Institute (ACCIS), Department of Aerospace Engineering, School of Civil, Aerospace, and Mechanical Engineering, University of Bristol, Bristol BS8 1TR, UK; [email protected] 3 Defence Science and Technology Laboratory, Porton Down, Salisbury SP4 0JQ, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-1483-686-248 Received: 6 March 2019; Accepted: 23 March 2019; Published: 28 March 2019 Abstract: The thermal decomposition of polyphenolic resins was studied by reactive molecular dynamics (RMD) simulation at elevated temperatures. Atomistic models of the polyphenolic resins to be used in the RMD were constructed using an automatic method which calls routines from the software package Materials Studio. In order to validate the models, simulated densities and heat capacities were compared with experimental values. The most suitable combination of force field and thermostat for this system was the Forcite force field with the Nosé–Hoover thermostat, which gave values of heat capacity closest to those of the experimental values. Simulated densities approached a final density of 1.05–1.08 g/cm3 which compared favorably with the experimental values of 1.16–1.21 g/cm3 for phenol-formaldehyde resins.