Analysis of Algae-Vulnerable Lakes in Utah Using R Plotting Tools to Visualize Water Quality Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

UMNP Mountains Manual 2017
Mountain Adventures Manual utahmasternaturalist.org June 2017 UMN/Manual/2017-03pr Welcome to Utah Master Naturalist! Utah Master Naturalist was developed to help you initiate or continue your own personal journey to increase your understanding of, and appreciation for, Utah’s amazing natural world. We will explore and learn aBout the major ecosystems of Utah, the plant and animal communities that depend upon those systems, and our role in shaping our past, in determining our future, and as stewards of the land. Utah Master Naturalist is a certification program developed By Utah State University Extension with the partnership of more than 25 other organizations in Utah. The mission of Utah Master Naturalist is to develop well-informed volunteers and professionals who provide education, outreach, and service promoting stewardship of natural resources within their communities. Our goal, then, is to assist you in assisting others to develop a greater appreciation and respect for Utah’s Beautiful natural world. “When we see the land as a community to which we belong, we may begin to use it with love and respect.” - Aldo Leopold Participating in a Utah Master Naturalist course provides each of us opportunities to learn not only from the instructors and guest speaKers, But also from each other. We each arrive at a Utah Master Naturalist course with our own rich collection of knowledge and experiences, and we have a unique opportunity to share that Knowledge with each other. This helps us learn and grow not just as individuals, but together as a group with the understanding that there is always more to learn, and more to share. -

Energy Loop Scenic Byway Corridor Management Plan Update Prepared By
ENERGY LOOP SCENIC BYWAY CORRIDOR MANAGEMENT PLAN UPDATE 2011 Energy Loop Scenic Byway Steering Committee: Jana Abrams, Energy Loop Scenic Byway Coordinator Bill Broadbear, US Forest Service Rosann Fillmore, US Forest Service Chanel Atwood, Castle Country Regional Information Center Tina Carter, Emery County Travel Bureau Kathy Hanna Smith, Carbon County Travel Bureau Kevin Christensen, Sanpete County Economic Development Mike McCandless, Emery County Economic Development Dan Richards, Utah State Parks Division Floyd Powell, Utah State Parks Division Nicole Nielson, Utah Division of Wildlife Resources Dale Stapley, Utah Department of Transportation Kevin Nichol, Utah Department of Transportation Gael Duffy Hill, Utah Office ofTourism PREPARED BY Fehr& Peers Bonneville Research 2180 South 1300 East Suite 220 170 South Main Street, Suite 775 Salt Lake City_ Utah 84106 Salt Lake City_ Utah 84101 p 801.463.7600 p 801.364.5300 "--' ""'-' CONTENTS '-.~· ""-' 1: Executive Summary Commercial Truck Traffic 35 1 ""'-" ~ 2: Introduction 5 6: Highway Safety and Management 35 "--" Byway Corridor Description 5 Commuter Traffic 36 ""-" ~ Purpose of Corridor Management Plan 6 Tourism Traffic 36 '<../ Guiding Purpose 9 Highway Safety Management Strategies 41 '-" Mission and Vision Statement 9 7: Interpretation 45 '-.../ "'-' Demographic Summary 49 3: Byway Organizational Plan 9 ""-" Goals 10 Energy Loop Key Travel/Tourism Information 49 ~ Byway Committee 11 Location and Access 49 """ "'-" Byway Coordinator 12 8: Demographics and Economic Development 49 -

Polar Bear Sightings at Antelope Island State Park- Water Temperatures at 27 Degrees
POLAR BEAR SIGHTINGS AT ANTELOPE ISLAND STATE PARK- WATER TEMPERATURES AT 27 DEGREES Utahns don crazy costumes and brave frigid air and water temperatures to raise funds for the Utah Special Olympics. DATE: Saturday, February 16 TIME: 10 a.m. LOCATION: Antelope Island State Park Marina Exit 332 off I-I5 Antelope Island State Park Assistant Manager Chris Haramoto reports water temperature at approximately 27 degrees. Due to salinity content, Great Salt Lake rarely freezes. Air temperature is expected to be near 40 degrees. Great Salt Lake State Marina hosts the 2008 Polar Plunge to benefit the Utah Special Olympics. Participants donate $25 for the privilege of jumping into the icy water, all to benefit a great cause. Wildlife viewing events for 2008 The DWR hosts several free wildlife-viewing events each year. The events provide a great opportunity for people to get outdoors and enjoy the state's wildlife! The events also provide great stories for the media and a chance to capture some awesome footage and photographs. More information is available in the latest Wildlife Review story titled "Get more than a glimpse -- Attend a Watchable Wildlife activity." Please click here to read the story: http://www.wildlife.utah.gov/wr/ Fishing volunteers still needed Training for adults who want to serve as volunteers in Utah's youth fishing clubs continues through mid-March. You can learn more by listening to the latest "Discover Utah Wildlife" radio shows. They're available at http://www.wildlife.utah.gov/radio/ . Big Game Hunters: You Can Still Apply for a Bonus Point or a Preference Point Applications accepted until Feb. -

2010 Utah Fishing Proclamation
Utah Division of Wildlife Resources • Turn in a poacher: 1-800-662-3337 • wildlife.utah.gov GUIDEBOOK FISHING 2010 UTAH 1 2010 • Fishing Utah For decades, CONTENTS Fishing a Utah fishing 2010 trip meant that 3 Contact information in Utah you would 3 Highlights bring home a stringer full of fat 5 General rules: licenses and rainbow trout. permits Today, you can still catch tasty 7 Fishing license fees rainbows, but you can also come 8 General rules: fishing methods 14 General rules: possession and Utah Fishing • Utah Fishing home with native cutthroats, walleye, striped bass, catfish, wipers and many transportation other species of fish. To learn more 16 Bag and possession limits 17 Fish consumption advisories about these fish, see the articles on 17 How to measure a fish pages 39–41. 18 Rules for specific waters Over the past year, there have 21 Community fishing waters been some exciting developments 33 Watercraft restrictions in the Division’s efforts to raise tiger 33 Utah’s boating laws and rules muskie here in Utah. You can read 35 Battling invasive species and about the past and future of this disease program in the article on page 41. 36 Did it get wet? Decontaminate it! You should also be aware of an 37 Catch-and-release fishing tips important regulation change that will 38 Restoring Utah’s rivers improve opportunity for all anglers at 39 Fish for something different Utah’s community fishing ponds. You’ll 40 A closer look at cutthroats find details in the article on page 46. 41 More tiger muskie for Utah Anglers of all ages and ability 42 Report illegal stocking levels find adventure in Utah’s diverse 43 Fishing facts fisheries. -

1992 Utah Fishing Proclamation
m ftroiG wm "t let erkLte^ "IHferae you won't let go ®fl Wfo(B ttrout! The largest fish ever taken on a rod and reel, a 3,427 pound great white shark, was caught on Berkley Trilene — America's best selling fishing Rtf*rlrlctir line! SPORTSCASTLE SANDY PRICE 5600 S. 9th E., Murray 838 E. 9400 S. 730 W. Prive River Rd. 263-3633 571-8812 637-2077 ZCMICENTER CEDAR CITY SUGARHOUSE 2nd Level ZCMI Center 606 S. Main 1171 East 2100 So.. 359-4540 586-0687 487-7726 VERNAL OGDEN CITY MALL ROY 872 W. Main 24th & Washington 5585 So. 1900 W. 789-0536 399-2310 776-4453 FAMILY CENTER PROVO/UNIV. MALL ROCK SPRINGS 5666 S. Redwood Rd. 1300 S. State 1371 Dewar Drive SPORTING GOODS COMPANY 967-9455 224-9115 307-362-4208 ON THE COVER "Snagged"by Luke Frazier, oil, 16"x CONTENTS 20". J99I, To learn more about Frazier and his art work, turn to page 60, INTRODUCTION Strawberry Recreation Area Loyal Clark, US Forest Service 35 One of the most exciting Director's Message Scofield Reservoir/What the Timothy H. Provan, Director Future Holds Kevin Christophereon, things about fishing is its Division of Wildlife Resources 2 unpredictability, You simply Southeast Region Fisheries Manager 37 Utah's 1992 Fishing Season don't kno w when that big Why Rainbow Trout? Bruce Schmidt, Fisheries Chief 3 Joe Valentine, Assistant Fisheries one is going to strike, it Chief (Culture) ........39 could be on your next cast! 1992 FISHING RULES Willard Bay Shad 1992 Fishing Rules: Purpose Thomas D. -

2016 Utah Angler Periodic Survey: Project Summary Report
Utah State University DigitalCommons@USU All In-stream Flows Material In-stream Flows 11-2017 2016 Utah Angler Periodic Survey: Project Summary Report R. J. Lilieholm Utah Division of Wildlife Resources J. M. Keating Utah Division of Wildlife Resources R. S. Krannich Utah Division of Wildlife Resources Follow this and additional works at: https://digitalcommons.usu.edu/instream_all Part of the Engineering Commons Recommended Citation Lilieholm, R. J.; Keating, J. M.; and Krannich, R. S., "2016 Utah Angler Periodic Survey: Project Summary Report" (2017). All In-stream Flows Material. Paper 10. https://digitalcommons.usu.edu/instream_all/10 This Report is brought to you for free and open access by the In-stream Flows at DigitalCommons@USU. It has been accepted for inclusion in All In-stream Flows Material by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. 2016 Utah Angler Periodic Survey Project Summary Report Prepared by R.J. Lilieholm, J.M. Keating, and R.S. Krannich Utah Division of Wildlife Resources November 2017 Table of Contents Executive Summary ...............................................................................................................iv Section 1: Introduction ...........................................................................................................1 Background and Justification ............................................................................................1 Building on Past Angler Surveys ......................................................................................2 -

RULES for SPECIFIC WATERS Utah Code § 23-20-3 and Utah Admin
RULES FOR SPECIFIC WATERS Utah Code § 23-20-3 and Utah Admin. Rule R657-13-20 The rules below take precedence over the general rules listed earlier in this guidebook. The seasons, bag limits and other restrictions in this section apply only to the waters listed below. General rules apply to all of the waters NOT listed in this section (see the Bag and Possession Limits section on page 16 to learn more about catching and harvesting fish at waters that are NOT listed in this section): American Fork Creek, Utah County From Utah Lake upstream to I-15. East from Utah Lake to I-15. • CLOSED March 1 through 6 a.m. on the first Saturday of May. Ashley Creek, Uintah County From Steinaker (Thornburg) diversion upstream to the water treatment plant near the mouth of Ashley Gorge. • Limit 2 trout. • ARTIFICIAL FLIES AND LURES ONLY. Aspen-Mirror Lake, Kane County • CLOSED Jan. 1 through 6 a.m. on the third Saturday of April. • Fishing from a boat or a float tube is unlawful. Badger Hollow, Wasatch County See Strawberry Reservoir tributaries. Barney Lake, Piute County • Limit 2 trout. • ARTIFICIAL FLIES AND LURES ONLY. Bear Lake, Rich County ▲ • See Fishing Across State Lines on pages 6–7 for license requirements. • Limit 2 trout. • Cutthroat trout or trout with cutthroat markings with all fins intact must be immediately released. Only cutthroat trout that have had one or more healed fins clipped may be kept. • Cisco may be taken with a handheld dipnet. Net opening may not exceed 18 inches in any dimension. -
U N S U U S E U R a C S
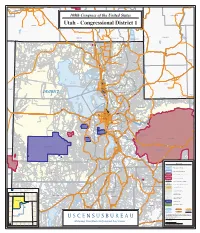
MINIDOKA Marbleton JEROME Heyburn Milner Lake Big Piney Burley CARIBOU Declo Murtaugh POWER TWIN FALLS BANNOCK Georgetown Downey SUBLETTE Albion 108th Congress of the United States r e v i R n e e Montpelier r G Malta Calpet CASSIA Oxford Oakley La Barge Paris Lower Goose Creek Reservoir Malad City Bloomington Clifton WYOMING Fontenelle Preston St. Charles Reservoir Dayton IDAHO Cokeville ONEIDA LINCOLN FRANKLIN Taylor Weston BEAR LAKE IDAHO IDAHO Franklin Fontenelle NEVADA UTAH Viva Naughton Reservoir Snowville Portage Cornish Lewiston Cove Garden Northwestern Shoshoni Res City Bear Lake 15 Clarkston Trenton S S t Richmond R 2 t 3 R t e te 84 1 89 Garden 4 Plymouth 2 Newton 91 Amalga Riverside StRte 30 (North Rd) Cache Smithfield Fielding S StRte 23 Laketown tH Hyde wy Diamondville 30 30 tRte Park S Bear Kemmerer Howell River North Oakley Peter Logan Opal Garland Logan River CACHE Benson Heights Tremonton Mendon Providence Dewey- ville Randolph SWEET- Millville Elwood WATER Honey- Nibley ville te 101 S tR RICH St Bear River Wellsville Hyrum Rte 83 City S t R S Granger t t e H 15 3 w 8 Paradise y 89 1 6 Avon Corinne Woodruff Brigham North Pass City S tHwy 3 t S 9 H w BOX ELDER Mantua y 1 6 Perry Carter Willard Neponset Reservoir South Willard Pleasant View Lyman Plain North Ogden WEBER Fort City Bridger Great Evanston Mountain View Harrisville Pineview Marriott- Utah Reservoir Salt Lake Farr General West te 39 Slaterville Dpo tR UINTA S n Cyn ) Huntsville de (Og West Haven Ogden Washington Terrace South Riverdale Ogden Robertson ELKO -

Board of the Governor's Office of Economic Development
Board of the Governor’s Office of Economic Development Governor’s Office of Economic Development Electronic Meeting – Zoom: https://us02web.zoom.us/j/83083749838?pwd=Tytma0pwY0RJV0RqVlEvM3JqM0Vldz09 By Phone: +16699006833,,83083749838# May 14, 2020 • 10:00 am – 12:00 pm AGENDA Welcome .................................................................................................... Mel Lavitt Motion on April 9, 2020 Meeting Minutes ............................................. GOED Board Incentives Report ....................................................................................... Mel Lavitt The Board will discuss public information about companies who have applied for incentives and vote on whether to approve the incentives, and if so, at what level. Three companies will be presented. Film Incentive Amendment................................................................ Virginia Pearce “Wireless” Office of Outdoor Recreation Grants ......................................................... Pitt Grewe 2002 Utah Children’s Outdoor Recreation & Education (UCORE) Grants 2002 Utah Outdoor Recreation (UORG) and Recreation Infrastructure (RRI) Grants GOED Update .............................................................................................. Val Hale Review of departmental activities and upcoming events Incentives Update ........................................................................... Tom Wadsworth Review of GOED’s new and existing corporate incentives projects EDCUtah Update ............................................................................. -

Utah State Parks Are Open Every Day Except for Thanksgiving and Christmas
PLAN YOUR TRIP Utah State Parks are open every day except for Thanksgiving and Christmas. For individual park hours visit our website stateparks.utah.gov. Full UTAH STATE PARK LOCATIONS / PARK RESERVATIONS 1 Anasazi AMENITIES Secure a campsite, pavilion, group area, or boat slip in advance by 2 Antelope Island calling 800-322-3770 8 a.m.–5 p.m. Monday through Friday, or visit 3 Bear Lake stateparks.utah.gov. # Center Visitor / Req. Fee Camping / Group Camping RV Sites Water Hookups—Partial Picnicking / Showers Restrooms Teepees / Yurts / Cabins / Fishing Boating / Biking Hiking Vehicles Off-Highway Golf / Zipline / Archery 84 Cache 3 State Parks 4 Camp Floyd Logan 1. Anasazi F-V R Reservations are always recommended. Individual campsite reservations 23 State Capitol Rivers 5 Coral Pink Sand Dunes Golden Spike Randolph N.H.S. Lakes 2. Antelope Island F-V C-G R-S B H-B may be made up to four months in advance and no fewer than two days Cities Box Elder Wasatch-Cashe N.F. 6 Dead Horse Point G Brigham City Rich 3. Bear Lake F-V C-G P-F R-S C B-F H-B before desired arrival date. Up to three individual campsite reservations per r e Interstate Highway 7 Deer Creek a 4. Camp Floyd Stagecoach Inn Museum F R t customer are permitted at most state parks. 43 U.S. Highway North S 8 East Canyon a 5. Coral Pink Sand Dunes F-V C-G P R-S H l Weber Morgan State Highway t PARK PASSES Ogden 9 Echo L 6. -

Deer Cove Pitch Book 011216
Table of Contents I Compelling Attributes .............. 1 II Deer Cove ....................... 5 III Jordanelle Specially Planned Area ..... 15 IV Residential Market ................. 21 V Hospitality Market ................. 26 VI Commercial Real Estate Overview ..... 33 VII Economic/Demographic Overview ..... 37 VIII Utah in the News .................. 46 IX Photos .......................... 59 Disclaimer: No representations or warranties, express or implied, are made with respect to the accuracy or completeness of the Information herein. The Information is subject to change and is not guaranteed as to completeness or accuracy. You understand that the Information is confidential and is furnished solely for the purpose of your review in connection with a potential investment in the property. COMPELLING ATTRIBUTES || I Opportunity Awaits World Class Mixed-Use Development Opportunity: The Deer Cove Master Planned Community (87 gross acres) represents one of the largest entitled mixed-use communities ready for development in the Park City/Deer Valley/Jordanelle Reservoir region. An infill location, with the closest proximity to existing on-mountain, freeway, and other public infrastructure, Deer Cove is located adjacent to U.S. 40, just off the Mayflower Interchange and within walking distance to both the Deer Crest Gondola and Jordanelle Reservoir. Deer Cove, with entitlements/approvals in-place, along with four on-mountain proposed luxury resorts developments will mark the beginning of the final build-out of the world-class Deer Valley/Deer Crest submarket. As such, with commanding mountain and lake views, from a mixed-use residential and hospitality perspective, Deer Cove is uniquely positioned to capture a broad spectrum of potential home buyers and hospitality patrons, who desire a value-oriented, year-round, contemporary alternative to the numerous near-by five star luxury on-mountain resorts and first generation homes located in the Park City/Deer Valley market. -

Utah Lake Watch Report 2009
Utah Lake Watch Report 2009 Utah State University Water Quality Extension Prepared by: Eric Peterson 1 Introduction As a statewide monitoring program, Utah Lake Watch (ULW) enlists the help of volunteers to collect data used to evaluate the general condition of Utah’ s lakes and reservoirs and how that changes over time. The data collected are used by the Utah Division of Water Quality and lake managers to determine whether the lake’s water quality is good enough to support the benefits that our lakes provide, such as recreational uses, fisheries and esthetic benefits. This report discusses the water transparency data (Secchi depths) and other related data collected in 2009 by citizen monitors throughout Utah. In 2009, USU’s Water Quality Extension program also conducted a pilot study on the use of citizen volunteers to collect bacterial data. The results of the pilot study will be reported in a separate document. Major objectives of the program include: •acquiring baseline data for Utah’s lakes and reservoirs; •providing education to state citizens on the importance of healthy lakes, how lakes function, and how to monitor lakes; and •demonstrating the effectiveness of citizen monitoring in collecting water quality data which can be used to better manage and protect our lakes and reservoirs To mee t these o bjecti ves parti ci pan ts are t rai ned to measure th e transparency an d make some simple observations for a particular lake or reservoir. Transparency correlates to other indicators of a lake’s condition, such as the amount of suspended algae growing in the lake, the amount of nutrients entering the lake, and the seasonal patterns of plant growth.