How Molecular Motors Extract Order from Chaos (A Key Issues Review)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
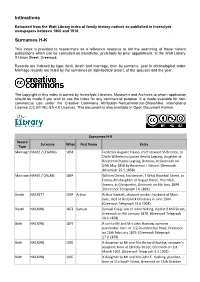
Intimations Surnames
Intimations Extracted from the Watt Library index of family history notices as published in Inverclyde newspapers between 1800 and 1918. Surnames H-K This index is provided to researchers as a reference resource to aid the searching of these historic publications which can be consulted on microfiche, preferably by prior appointment, at the Watt Library, 9 Union Street, Greenock. Records are indexed by type: birth, death and marriage, then by surname, year in chronological order. Marriage records are listed by the surnames (in alphabetical order), of the spouses and the year. The copyright in this index is owned by Inverclyde Libraries, Museums and Archives to whom application should be made if you wish to use the index for any commercial purpose. It is made available for non- commercial use under the Creative Commons Attribution-Noncommercial-ShareAlike International License (CC BY-NC-SA 4.0 License). This document is also available in Open Document Format. Surnames H-K Record Surname When First Name Entry Type Marriage HAASE / LEGRING 1858 Frederick Auguste Haase, chief steward SS Bremen, to Ottile Wilhelmina Louise Amelia Legring, daughter of Reverend Charles Legring, Bremen, at Greenock on 24th May 1858 by Reverend J. Nelson. (Greenock Advertiser 25.5.1858) Marriage HAASE / OHLMS 1894 William Ohlms, hairdresser, 7 West Blackhall Street, to Emma, 4th daughter of August Haase, Herrnhut, Saxony, at Glengarden, Greenock on 6th June 1894 .(Greenock Telegraph 7.6.1894) Death HACKETT 1904 Arthur Arthur Hackett, shipyard worker, husband of Mary Jane, died at Greenock Infirmary in June 1904. (Greenock Telegraph 13.6.1904) Death HACKING 1878 Samuel Samuel Craig, son of John Hacking, died at 9 Mill Street, Greenock on 9th January 1878. -

Chloroplast Transit Peptides: Structure, Function and Evolution
reviews Chloroplast transit Although the first demonstration of precursor trans- port into chloroplasts was shown over two decades peptides: structure, ago3,4, only now is this area of cell biology becom- ing well understood. Many excellent reviews have been published recently on the evolution of plas- function and tids5, the evolution of organelle genomes6, the mechanism of gene transfer from organelles to the nucleus7 and the mechanism of protein import into evolution chloroplasts8,9. Proteins destined to plastids and other organ- elles share in common the requirement for ‘new’ Barry D. Bruce sequence information to facilitate their correct trafficking within the cell. Although in most cases this information resides in a cleavable, N-terminal sequence often collectively referred to as signal It is thought that two to three thousand different proteins are sequence, the different organelle-targeting se- targeted to the chloroplast, and the ‘transit peptides’ that act as quences have distinct properties and names: ‘signal peptides’ for the endoplasmic reticulum, chloroplast targeting sequences are probably the largest class of ‘presequences’ for the mitochondria and ‘transit peptides’ for chloroplasts and other plastids. This targeting sequences in plants. At a primary structural level, transit review focuses on recent progress in dissecting peptide sequences are highly divergent in length, composition and the role of the stromal-targeting domain of chloro- plast transit peptides. I will consider briefly the organization. An emerging concept suggests that transit peptides multitude of distinct functions that transit peptides contain multiple domains that provide either distinct or overlapping perform, provide an update on the limited struc- tural information of a number of transit peptides functions. -

JM Coetzee and Mathematics Peter Johnston
1 'Presences of the Infinite': J. M. Coetzee and Mathematics Peter Johnston PhD Royal Holloway University of London 2 Declaration of Authorship I, Peter Johnston, hereby declare that this thesis and the work presented in it is entirely my own. Where I have consulted the work of others, this is always clearly stated. Signed: Dated: 3 Abstract This thesis articulates the resonances between J. M. Coetzee's lifelong engagement with mathematics and his practice as a novelist, critic, and poet. Though the critical discourse surrounding Coetzee's literary work continues to flourish, and though the basic details of his background in mathematics are now widely acknowledged, his inheritance from that background has not yet been the subject of a comprehensive and mathematically- literate account. In providing such an account, I propose that these two strands of his intellectual trajectory not only developed in parallel, but together engendered several of the characteristic qualities of his finest work. The structure of the thesis is essentially thematic, but is also broadly chronological. Chapter 1 focuses on Coetzee's poetry, charting the increasing involvement of mathematical concepts and methods in his practice and poetics between 1958 and 1979. Chapter 2 situates his master's thesis alongside archival materials from the early stages of his academic career, and thus traces the development of his philosophical interest in the migration of quantificatory metaphors into other conceptual domains. Concentrating on his doctoral thesis and a series of contemporaneous reviews, essays, and lecture notes, Chapter 3 details the calculated ambivalence with which he therein articulates, adopts, and challenges various statistical methods designed to disclose objective truth. -

Construction and Loss of Bacterial Flagellar Filaments
biomolecules Review Construction and Loss of Bacterial Flagellar Filaments Xiang-Yu Zhuang and Chien-Jung Lo * Department of Physics and Graduate Institute of Biophysics, National Central University, Taoyuan City 32001, Taiwan; [email protected] * Correspondence: [email protected] Received: 31 July 2020; Accepted: 4 November 2020; Published: 9 November 2020 Abstract: The bacterial flagellar filament is an extracellular tubular protein structure that acts as a propeller for bacterial swimming motility. It is connected to the membrane-anchored rotary bacterial flagellar motor through a short hook. The bacterial flagellar filament consists of approximately 20,000 flagellins and can be several micrometers long. In this article, we reviewed the experimental works and models of flagellar filament construction and the recent findings of flagellar filament ejection during the cell cycle. The length-dependent decay of flagellar filament growth data supports the injection-diffusion model. The decay of flagellar growth rate is due to reduced transportation of long-distance diffusion and jamming. However, the filament is not a permeant structure. Several bacterial species actively abandon their flagella under starvation. Flagellum is disassembled when the rod is broken, resulting in an ejection of the filament with a partial rod and hook. The inner membrane component is then diffused on the membrane before further breakdown. These new findings open a new field of bacterial macro-molecule assembly, disassembly, and signal transduction. Keywords: self-assembly; injection-diffusion model; flagellar ejection 1. Introduction Since Antonie van Leeuwenhoek observed animalcules by using his single-lens microscope in the 18th century, we have entered a new era of microbiology. -

Review of Molecular Motors
REVIEWS CYTOSKELETAL MOTORS Moving into the cell: single-molecule studies of molecular motors in complex environments Claudia Veigel*‡ and Christoph F. Schmidt§ Abstract | Much has been learned in the past decades about molecular force generation. Single-molecule techniques, such as atomic force microscopy, single-molecule fluorescence microscopy and optical tweezers, have been key in resolving the mechanisms behind the power strokes, ‘processive’ steps and forces of cytoskeletal motors. However, it remains unclear how single force generators are integrated into composite mechanical machines in cells to generate complex functions such as mitosis, locomotion, intracellular transport or mechanical sensory transduction. Using dynamic single-molecule techniques to track, manipulate and probe cytoskeletal motor proteins will be crucial in providing new insights. Molecular motors are machines that convert free energy, data suggest that during the force-generating confor- mostly obtained from ATP hydrolysis, into mechanical mational change, known as the power stroke, the lever work. The cytoskeletal motor proteins of the myosin and arm of myosins8,11 rotates around its base at the catalytic kinesin families, which interact with actin filaments and domain11–17, which can cause the displacement of bound microtubules, respectively, are the best understood. Less cargo by several nanometres18 (FIG. 1B). In kinesins, the is known about the dynein family of cytoskeletal motors, switching of the neck linker (~13 amino acids connecting which interact with microtubules. Cytoskeletal motors the catalytic core to the cargo-binding stalk domain) from power diverse forms of motility, ranging from the move- an ‘undocked’ state to a state in which it is ‘docked’ to the ment of entire cells (as occurs in muscular contraction catalytic domain, is the equivalent of the myosin power or cell locomotion) to intracellular structural dynamics stroke10. -

Control of Molecular Motor Motility in Electrical Devices
Control of Molecular Motor Motility in Electrical Devices Thesis submitted in accordance with the requirements of The University of Liverpool for the degree of Doctor in Philosophy By Laurence Charles Ramsey Department of Electrical Engineering & Electronics April 2014 i Abstract In the last decade there has been increased interest in the study of molecular motors. Motor proteins in particular have gained a large following due to their high efficiency of force generation and the ability to incorporate the motors into linear device designs. Much of the recent research centres on using these protein systems to transport cargo around the surface of a device. The studies carried out in this thesis aim to investigate the use of molecular motors in lab- on-a-chip devices. Two distinct motor protein systems are used to show the viability of utilising these nanoscale machines as a highly specific and controllable method of transporting molecules around the surface of a lab-on-a-chip device. Improved reaction kinetics and increased detection sensitivity are just two advantages that could be achieved if a motor protein system could be incorporated and appropriately controlled within a device such as an immunoassay or microarray technologies. The first study focuses on the motor protein system Kinesin. This highly processive motor is able to propel microtubules across a surface and has shown promise as an in vitro nanoscale transport system. A novel device design is presented where the motility of microtubules is controlled using the combination of a structured surface and a thermoresponsive polymer. Both topographic confinement of the motility and the creation of localised ‘gates’ are used to show a method for the control and guidance of microtubules. -

On Helicases and Other Motor Proteins Eric J Enemark and Leemor Joshua-Tor
Available online at www.sciencedirect.com On helicases and other motor proteins Eric J Enemark and Leemor Joshua-Tor Helicases are molecular machines that utilize energy derived to carry out the repetitive mechanical operation of prying from ATP hydrolysis to move along nucleic acids and to open base pairs and/or actively translocating with respect separate base-paired nucleotides. The movement of the to the nucleic acid substrate. Many other molecular helicase can also be described as a stationary helicase that motors utilize similar engines to carry out multiple pumps nucleic acid. Recent structural data for the hexameric diverse functions such as translocation of peptides E1 helicase of papillomavirus in complex with single-stranded in the case of ClpX, movement along cellular structures DNA and MgADP has provided a detailed atomic and in the case of dyenin, and rotation about an axle as in mechanistic picture of its ATP-driven DNA translocation. The F1-ATPase. structural and mechanistic features of this helicase are compared with the hexameric helicase prototypes T7gp4 and Based upon conserved sequence motifs, helicases have SV40 T-antigen. The ATP-binding site architectures of these been classified into six superfamilies [1,2]. An extensive proteins are structurally similar to the sites of other prototypical review of these superfamilies has been provided recently ATP-driven motors such as F1-ATPase, suggesting related [3]. Superfamily 1 (SF1) and superfamily 2 (SF2) heli- roles for the individual site residues in the ATPase activity. cases are very prevalent, generally monomeric, and participate in several diverse DNA and RNA manipula- Address tions. -

Cytoskeleton and Cell Motility
Cytoskeleton and Cell Motility Thomas Risler1 1Laboratoire Physicochimie Curie (CNRS-UMR 168), Institut Curie, Section de Recherche, 26 rue d’Ulm, 75248 Paris Cedex 05, France (Dated: January 23, 2008) PACS numbers: 1 Article Outline Glossary I. Definition of the Subject and Its Importance II. Introduction III. The Diversity of Cell Motility A. Swimming B. Crawling C. Extensions of cell motility IV. The Cell Cytoskeleton A. Biopolymers B. Molecular motors C. Motor families D. Other cytoskeleton-associated proteins E. Cell anchoring and regulatory pathways F. The prokaryotic cytoskeleton V. Filament-Driven Motility A. Microtubule growth and catastrophes B. Actin gels C. Modeling polymerization forces D. A model system for studying actin-based motility: The bacterium Listeria mono- cytogenes E. Another example of filament-driven amoeboid motility: The nematode sperm cell VI. Motor-Driven Motility A. Generic considerations 2 B. Phenomenological description close to thermodynamic equilibrium C. Hopping and transport models D. The two-state model E. Coupled motors and spontaneous oscillations F. Axonemal beating VII. Putting It Together: Active Polymer Solutions A. Mesoscopic approaches B. Microscopic approaches C. Macroscopic phenomenological approaches: The active gels D. Comparisons of the different approaches to describing active polymer solutions VIII. Extensions and Future Directions Acknowledgments Bibliography Glossary Cell Structural and functional elementary unit of all life forms. The cell is the smallest unit that can be characterized as living. Eukaryotic cell Cell that possesses a nucleus, a small membrane-bounded compartment that contains the genetic material of the cell. Cells that lack a nucleus are called prokaryotic cells or prokaryotes. Domains of life archaea, bacteria and eukarya - or in English eukaryotes, and made of eukaryotic cells - which constitute the three fundamental branches in which all life forms are classified. -

The Fessard's School of Neurophysiology After
The fessard’s School of neurophysiology after the Second World War in france: globalisation and diversity in neurophysiological research (1938-1955) Jean-Gaël Barbara To cite this version: Jean-Gaël Barbara. The fessard’s School of neurophysiology after the Second World War in france: globalisation and diversity in neurophysiological research (1938-1955). Archives Italiennes de Biologie, Universita degli Studi di Pisa, 2011. halshs-03090650 HAL Id: halshs-03090650 https://halshs.archives-ouvertes.fr/halshs-03090650 Submitted on 11 Jan 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. The Fessard’s School of neurophysiology after the Second World War in France: globalization and diversity in neurophysiological research (1938-1955) Jean- Gaël Barbara Université Pierre et Marie Curie, Paris, Centre National de la Recherche Scientifique, CNRS UMR 7102 Université Denis Diderot, Paris, Centre National de la Recherche Scientifique, CNRS UMR 7219 [email protected] Postal Address : JG Barbara, UPMC, case 14, 7 quai Saint Bernard, 75005, -

Missouri State Archives Finding Aid 5.20
Missouri State Archives Finding Aid 5.20 OFFICE OF SECRETARY OF STATE COMMISSIONS PARDONS, 1836- Abstract: Pardons (1836-2018), restorations of citizenship, and commutations for Missouri convicts. Extent: 66 cubic ft. (165 legal-size Hollinger boxes) Physical Description: Paper Location: MSA Stacks ADMINISTRATIVE INFORMATION Alternative Formats: Microfilm (S95-S123) of the Pardon Papers, 1837-1909, was made before additions, interfiles, and merging of the series. Most of the unmicrofilmed material will be found from 1854-1876 (pardon certificates and presidential pardons from an unprocessed box) and 1892-1909 (formerly restorations of citizenship). Also, stray records found in the Senior Reference Archivist’s office from 1836-1920 in Box 164 and interfiles (bulk 1860) from 2 Hollinger boxes found in the stacks, a portion of which are in Box 164. Access Restrictions: Applications or petitions listing the social security numbers of living people are confidential and must be provided to patrons in an alternative format. At the discretion of the Senior Reference Archivist, some records from the Board of Probation and Parole may be restricted per RSMo 549.500. Publication Restrictions: Copyright is in the public domain. Preferred Citation: [Name], [Date]; Pardons, 1836- ; Commissions; Office of Secretary of State, Record Group 5; Missouri State Archives, Jefferson City. Acquisition Information: Agency transfer. PARDONS Processing Information: Processing done by various staff members and completed by Mary Kay Coker on October 30, 2007. Combined the series Pardon Papers and Restorations of Citizenship because the latter, especially in later years, contained a large proportion of pardons. The two series were split at 1910 but a later addition overlapped from 1892 to 1909 and these records were left in their respective boxes but listed chronologically in the finding aid. -

1. Overview 2. Database Architecture 3. Example Tree 6. Mentorship Network Influences?
Neurotree: Graphing the academic genealogy of neuroscience Stephen V. David1, Will Chertoff1, Titipat Achakulvisut2, Daniel Acuna2 1Oregon Health & Science University, 2Northwestern University 1. Overview 3. Example tree 4. Founding ancestors Neurotree (http://neurotree.org, [1]) is a collaborative, open-access Name N Resarch area Family tree The distance between two nodes can be Johannes Müller 7715 Physiology website that tracks and visualizes the academic genealogy and history P+ William Fitch measured by the number of mentorship Sir Charles Sherrington 4758 Neurophysiology of neuroscience. After 10 years of growth driven by user-generated Allen Hermann von Helmholtz 3048 Psychophysics University of P- steps connecting them through a Sir John Eccles 2998 Synapses content, the site has captured information about the mentorship of over Rudolf Oregon Ludwig Robert Samuel Alexander Sir Charles Medical common ancestor i(below). The list at Karl Lashley 2558 Learning and memory 80,000 neuroscientists. It has become a unique tool for a community of John Friedrich Karl Koch Sir Charles Kinnier Charles Gordon John Scott Sir Charles Harvey Sir Charles Karl Edgar School C+ Louis Agassiz 2241 Anatomy Sir Michael Newport Goltz Virchow Universität Scott Wilson Symonds Holmes Farquhar Sherrington Scott Williams Scott Spencer Wilder Douglas Frederic right shows the 30 most frequent primary researchers, students, journal editors, and the press. Once Foster Langley Kaiser-Wilhelms- Universität Berlin (ID Sherrington National Hospital, Queen National -
![Arxiv:1105.2423V1 [Physics.Bio-Ph] 12 May 2011 C](https://docslib.b-cdn.net/cover/6992/arxiv-1105-2423v1-physics-bio-ph-12-may-2011-c-1406992.webp)
Arxiv:1105.2423V1 [Physics.Bio-Ph] 12 May 2011 C
Cytoskeleton and Cell Motility Thomas Risler Institut Curie, Centre de Recherche, UMR 168 (UPMC Univ Paris 06, CNRS), 26 rue d'Ulm, F-75005 Paris, France Article Outline C. Macroscopic phenomenological approaches: The active gels Glossary D. Comparisons of the different approaches to de- scribing active polymer solutions I. Definition of the Subject and Its Importance VIII. Extensions and Future Directions II. Introduction Acknowledgments III. The Diversity of Cell Motility Bibliography A. Swimming B. Crawling C. Extensions of cell motility IV. The Cell Cytoskeleton A. Biopolymers B. Molecular motors C. Motor families D. Other cytoskeleton-associated proteins E. Cell anchoring and regulatory pathways F. The prokaryotic cytoskeleton V. Filament-Driven Motility A. Microtubule growth and catastrophes B. Actin gels C. Modeling polymerization forces D. A model system for studying actin-based motil- ity: The bacterium Listeria monocytogenes E. Another example of filament-driven amoeboid motility: The nematode sperm cell VI. Motor-Driven Motility A. Generic considerations B. Phenomenological description close to thermo- dynamic equilibrium arXiv:1105.2423v1 [physics.bio-ph] 12 May 2011 C. Hopping and transport models D. The two-state model E. Coupled motors and spontaneous oscillations F. Axonemal beating VII. Putting It Together: Active Polymer Solu- tions A. Mesoscopic approaches B. Microscopic approaches 2 Glossary I. DEFINITION OF THE SUBJECT AND ITS IMPORTANCE Cell Structural and functional elementary unit of all life forms. The cell is the smallest unit that can be We, as human beings, are made of a collection of cells, characterized as living. which are most commonly considered as the elementary building blocks of all living forms on earth [1].