Spiralian Phylogeny Informs the Evolution of Microscopic Lineages
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lesson 4 Plant and Animal Classification and Identification
LESSON 4 PLANT AND ANIMAL CLASSIFICATION AND IDENTIFICATION Aim Become familiar with the basic ways in which plants and animals can be classified and identified CLASSIFICATION OF ORGANISMS Living organisms that are relevant to the ecotourism tour guide are generally divided into two main groups of things: ANIMALS........and..........PLANTS The boundaries between these two groups are not always clear, and there is an increasing tendency to recognise as many as five or six kingdoms. However, for our purposes we will stick with the two kingdoms. Each of these main groups is divided up into a number of smaller groups called 'PHYLA' (or Phylum). Each phylum has certain broad characteristics which distinguish it from other groups of plants or animals. For example, all flowering plants are in the phyla 'Anthophyta' (if a plant has a flower it is a member of the phyla 'Anthophyta'). All animals which have a backbone are members of the phyla 'Vertebrate' (i.e. Vertebrates). Getting to know these major groups of plants and animals is the first step in learning the characteristics of, and learning to identify living things. You should try to gain an understanding of the order of the phyla - from the simplest to the most complex - in both the animal and plant kingdoms. Gaining this type of perspective will provide you with a framework which you can slot plants or animals into when trying to identify them in nature. Understanding the phyla is only the first step, however. Phyla are further divided into groups called 'classes'. Classes are divided and divided again and so on. -

Systema Naturae∗
Systema Naturae∗ c Alexey B. Shipunov v. 5.802 (June 29, 2008) 7 Regnum Monera [ Bacillus ] /Bacteria Subregnum Bacteria [ 6:8Bacillus ]1 Superphylum Posibacteria [ 6:2Bacillus ] stat.m. Phylum 1. Firmicutes [ 6Bacillus ]2 Classis 1(1). Thermotogae [ 5Thermotoga ] i.s. 2(2). Mollicutes [ 5Mycoplasma ] 3(3). Clostridia [ 5Clostridium ]3 4(4). Bacilli [ 5Bacillus ] 5(5). Symbiobacteres [ 5Symbiobacterium ] Phylum 2. Actinobacteria [ 6Actynomyces ] Classis 1(6). Actinobacteres [ 5Actinomyces ] Phylum 3. Hadobacteria [ 6Deinococcus ] sed.m. Classis 1(7). Hadobacteres [ 5Deinococcus ]4 Superphylum Negibacteria [ 6:2Rhodospirillum ] stat.m. Phylum 4. Chlorobacteria [ 6Chloroflexus ]5 Classis 1(8). Ktedonobacteres [ 5Ktedonobacter ] sed.m. 2(9). Thermomicrobia [ 5Thermomicrobium ] 3(10). Chloroflexi [ 5Chloroflexus ] ∗Only recent taxa. Viruses are not included. Abbreviations and signs: sed.m. (sedis mutabilis); stat.m. (status mutabilis): s., aut i. (superior, aut interior); i.s. (incertae sedis); sed.p. (sedis possibilis); s.str. (sensu stricto); s.l. (sensu lato); incl. (inclusum); excl. (exclusum); \quotes" for environmental groups; * (asterisk) for paraphyletic taxa; / (slash) at margins for major clades (\domains"). 1Incl. \Nanobacteria" i.s. et dubitativa, \OP11 group" i.s. 2Incl. \TM7" i.s., \OP9", \OP10". 3Incl. Dictyoglomi sed.m., Fusobacteria, Thermolithobacteria. 4= Deinococcus{Thermus. 5Incl. Thermobaculum i.s. 1 4(11). Dehalococcoidetes [ 5Dehalococcoides ] 5(12). Anaerolineae [ 5Anaerolinea ]6 Phylum 5. Cyanobacteria [ 6Nostoc ] Classis 1(13). Gloeobacteres [ 5Gloeobacter ] 2(14). Chroobacteres [ 5Chroococcus ]7 3(15). Hormogoneae [ 5Nostoc ] Phylum 6. Bacteroidobacteria [ 6Bacteroides ]8 Classis 1(16). Fibrobacteres [ 5Fibrobacter ] 2(17). Chlorobi [ 5Chlorobium ] 3(18). Salinibacteres [ 5Salinibacter ] 4(19). Bacteroidetes [ 5Bacteroides ]9 Phylum 7. Spirobacteria [ 6Spirochaeta ] Classis 1(20). Spirochaetes [ 5Spirochaeta ] s.l.10 Phylum 8. Planctobacteria [ 6Planctomyces ]11 Classis 1(21). -

Platyhelminthes, Nemertea, and "Aschelminthes" - A
BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. III - Platyhelminthes, Nemertea, and "Aschelminthes" - A. Schmidt-Rhaesa PLATYHELMINTHES, NEMERTEA, AND “ASCHELMINTHES” A. Schmidt-Rhaesa University of Bielefeld, Germany Keywords: Platyhelminthes, Nemertea, Gnathifera, Gnathostomulida, Micrognathozoa, Rotifera, Acanthocephala, Cycliophora, Nemathelminthes, Gastrotricha, Nematoda, Nematomorpha, Priapulida, Kinorhyncha, Loricifera Contents 1. Introduction 2. General Morphology 3. Platyhelminthes, the Flatworms 4. Nemertea (Nemertini), the Ribbon Worms 5. “Aschelminthes” 5.1. Gnathifera 5.1.1. Gnathostomulida 5.1.2. Micrognathozoa (Limnognathia maerski) 5.1.3. Rotifera 5.1.4. Acanthocephala 5.1.5. Cycliophora (Symbion pandora) 5.2. Nemathelminthes 5.2.1. Gastrotricha 5.2.2. Nematoda, the Roundworms 5.2.3. Nematomorpha, the Horsehair Worms 5.2.4. Priapulida 5.2.5. Kinorhyncha 5.2.6. Loricifera Acknowledgements Glossary Bibliography Biographical Sketch Summary UNESCO – EOLSS This chapter provides information on several basal bilaterian groups: flatworms, nemerteans, Gnathifera,SAMPLE and Nemathelminthes. CHAPTERS These include species-rich taxa such as Nematoda and Platyhelminthes, and as taxa with few or even only one species, such as Micrognathozoa (Limnognathia maerski) and Cycliophora (Symbion pandora). All Acanthocephala and subgroups of Platyhelminthes and Nematoda, are parasites that often exhibit complex life cycles. Most of the taxa described are marine, but some have also invaded freshwater or the terrestrial environment. “Aschelminthes” are not a natural group, instead, two taxa have been recognized that were earlier summarized under this name. Gnathifera include taxa with a conspicuous jaw apparatus such as Gnathostomulida, Micrognathozoa, and Rotifera. Although they do not possess a jaw apparatus, Acanthocephala also belong to Gnathifera due to their epidermal structure. ©Encyclopedia of Life Support Systems (EOLSS) BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. -

Systematics , Zoogeography , Andecologyofthepriapu
SYSTEMATICS, ZOOGEOGRAPHY, AND ECOLOGY OF THE PRIAPULIDA by J. VAN DER LAND (Rijksmuseum van Natuurlijke Historie, Leiden) With 89 text-figures and five plates CONTENTS Ι Introduction 4 1.1 Plan 4 1.2 Material and methods 5 1.3 Acknowledgements 6 2 Systematics 8 2.1 Historical introduction 8 2.2 Phylum Priapulida 9 2.2.1 Diagnosis 10 2.2.2 Classification 10 2.2.3 Definition of terms 12 2.2.4 Basic adaptive features 13 2.2.5 General features 13 2.2.6 Systematic notes 2 5 2.3 Key to the taxa 20 2.4 Survey of the taxa 29 Priapulidae 2 9 Priapulopsis 30 Priapulus 51 Acanthopriapulus 60 Halicryptus 02 Tubiluchidae 7° Tubiluchus 73 3 Zoogeography and ecology 84 3.1 Distribution 84 3.1.1 Faunistics 90 3.1.2 Expeditions 93 3.2 Ecology 96 3.2.1 Substratum 96 3.2.2 Depth 97 3.2.3 Temperature 97 3.24 Salinity 98 3.2.5 Oxygen 98 3.2.6 Food 99 3.2.7 Predators 100 3.2.8 Communities 102 3.3 Theoretical zoogeography 102 3.3.1 Bipolarity 102 3.3.2 Relict distribution 103 3.3.3 Concluding remarks 104 References 105 4 ZOOLOGISCHE VERHANDELINGEN 112 (1970) 1 INTRODUCTION The phylum Priapulida is only a very small group of marine worms but, since these animals apparently represent the last remnants of a once un- doubtedly much more important animal type, they are certainly of great scientific interest. Therefore, it is to be regretted that a comprehensive review of our present knowledge of the group does not exist, which does not only cause the faulty way in which the Priapulida are treated usually in textbooks and works on phylogeny, but also hampers further studies. -

Phylum Nemertea)
THE BIOLOGY AND SYSTEMATICS OF A NEW SPECIES OF RIBBON WORM, GENUS TUBULANUS (PHYLUM NEMERTEA) By Rebecca Kirk Ritger Submitted to the Faculty of the College of Arts and Sciences of American University in Partial Fulfillment of the Requirements for the Degree of Master of Science In Biology Chair: Dr. Qiristopher'Tudge m Dr.David C r. Jon L. Norenburg Dean of the College of Arts and Sciences JuK4£ __________ Date 2004 American University Washington, D.C. 20016 AMERICAN UNIVERSITY LIBRARY 1 1 0 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. UMI Number: 1421360 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. ® UMI UMI Microform 1421360 Copyright 2004 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. Box 1346 Ann Arbor, Ml 48106-1346 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. THE BIOLOGY AND SYSTEMATICS OF A NEW SPECIES OF RIBBON WORM, GENUS TUBULANUS (PHYLUM NEMERTEA) By Rebecca Kirk Ritger ABSTRACT Most nemerteans are studied from poorly preserved museum specimens. -

Defining Phyla: Evolutionary Pathways to Metazoan Body Plans
EVOLUTION & DEVELOPMENT 3:6, 432-442 (2001) Defining phyla: evolutionary pathways to metazoan body plans Allen G. Collins^ and James W. Valentine* Museum of Paleontology and Department of Integrative Biology, University of California, Berkeley, CA 94720, USA 'Author for correspondence (email: [email protected]) 'Present address: Section of Ecology, Befiavior, and Evolution, Division of Biology, University of California, San Diego, La Jolla, CA 92093-0116, USA SUMMARY Phyla are defined by two sets of criteria, one pothesis of Nielsen; the clonal hypothesis of Dewel; the set- morphological and the other historical. Molecular evidence aside cell hypothesis of Davidson et al.; and a benthic hy- permits the grouping of animals into clades and suggests that pothesis suggested by the fossil record. It is concluded that a some groups widely recognized as phyla are paraphyletic, benthic radiation of animals could have supplied the ances- while some may be polyphyletic; the phyletic status of crown tral lineages of all but a few phyla, is consistent with molecu- phyla is tabulated. Four recent evolutionary scenarios for the lar evidence, accords well with fossil evidence, and accounts origins of metazoan phyla and of supraphyletic clades are as- for some of the difficulties in phylogenetic analyses of phyla sessed in the light of a molecular phylogeny: the trochaea hy- based on morphological criteria. INTRODUCTION Molecules have provided an important operational ad- vance to addressing questions about the origins of animal Concepts of animal phyla have changed importantly from phyla. Molecular developmental and comparative genomic their origins in the six Linnaean classis and four Cuvieran evidence offer insights into the genetic bases of body plan embranchements. -

Convergent Evolution of the Ladder-Like Ventral Nerve Cord in Annelida Conrad Helm1*, Patrick Beckers2, Thomas Bartolomaeus2, Stephan H
Helm et al. Frontiers in Zoology (2018) 15:36 https://doi.org/10.1186/s12983-018-0280-y RESEARCH Open Access Convergent evolution of the ladder-like ventral nerve cord in Annelida Conrad Helm1*, Patrick Beckers2, Thomas Bartolomaeus2, Stephan H. Drukewitz3, Ioannis Kourtesis1, Anne Weigert4, Günter Purschke5, Katrine Worsaae6, Torsten H. Struck7 and Christoph Bleidorn1,8* Abstract Background: A median, segmented, annelid nerve cord has repeatedly been compared to the arthropod and vertebrate nerve cords and became the most used textbook representation of the annelid nervous system. Recent phylogenomic analyses, however, challenge the hypothesis that a subepidermal rope-ladder-like ventral nerve cord (VNC) composed of a paired serial chain of ganglia and somata-free connectives represents either a plesiomorphic or a typical condition in annelids. Results: Using a comparative approach by combining phylogenomic analyses with morphological methods (immunohistochemistry and CLSM, histology and TEM), we compiled a comprehensive dataset to reconstruct the evolution of the annelid VNC. Our phylogenomic analyses generally support previous topologies. However, the so far hard-to-place Apistobranchidae and Psammodrilidae are now incorporated among the basally branching annelids with high support. Based on this topology we reconstruct an intraepidermal VNC as the ancestral state in Annelida. Thus, a subepidermal ladder-like nerve cord clearly represents a derived condition. Conclusions: Based on the presented data, a ladder-like appearance of the ventral nerve cord evolved repeatedly, and independently of the transition from an intraepidermal to a subepidermal cord during annelid evolution. Our investigations thereby propose an alternative set of neuroanatomical characteristics for the last common ancestor of Annelida or perhaps even Spiralia. -

Xenacoelomorpha's Significance for Understanding Bilaterian Evolution
Available online at www.sciencedirect.com ScienceDirect Xenacoelomorpha’s significance for understanding bilaterian evolution Andreas Hejnol and Kevin Pang The Xenacoelomorpha, with its phylogenetic position as sister biology models are the fruitfly Drosophila melanogaster and group of the Nephrozoa (Protostomia + Deuterostomia), plays the nematode Caenorhabditis elegans, in which basic prin- a key-role in understanding the evolution of bilaterian cell types ciples of developmental processes have been studied in and organ systems. Current studies of the morphological and great detail. It might be because the field of evolutionary developmental diversity of this group allow us to trace the developmental biology — EvoDevo — has its origin in evolution of different organ systems within the group and to developmental biology and not evolutionary biology that reconstruct characters of the most recent common ancestor of species under investigation are often called ‘model spe- Xenacoelomorpha. The disparity of the clade shows that there cies’. Criteria for selected representative species are cannot be a single xenacoelomorph ‘model’ species and primarily the ease of access to collected material and strategic sampling is essential for understanding the evolution their ability to be cultivated in the lab [1]. In some cases, of major traits. With this strategy, fundamental insights into the a supposedly larger number of ancestral characters or a evolution of molecular mechanisms and their role in shaping dominant role in ecosystems have played an additional animal organ systems can be expected in the near future. role in selecting model species. These arguments were Address used to attract sufficient funding for genome sequencing Sars International Centre for Marine Molecular Biology, University of and developmental studies that are cost-intensive inves- Bergen, Thormøhlensgate 55, 5008 Bergen, Norway tigations. -

Invertebrates Invertebrates: • Are Animals Without Backbones • Represent 95% of the Animal Kingdom Animal Diversity Morphological Vs
Invertebrates Invertebrates: • Are animals without backbones • Represent 95% of the animal kingdom Animal Diversity Morphological vs. Molecular Character Phylogeny? A tree is a hypothesis supported or not supported by evidence. Groupings change as new evidence become available. Sponges - Porifera Natural Bath Sponges – over-collected, now uncommon Sponges • Perhaps oldest animal phylum (Ctenphora possibly older) • may represent several old phyla, some now extinct ----------------Ctenophora? Sponges - Porifera • Mostly marine • Sessile animals • Lack true tissues; • Have only a few cell types, cells kind of independent • Most have no symmetry • Body resembles a sac perforated with holes, system of canals. • Strengthened by fibers of spongin, spicules Sponges have a variety of shapes Sponges Pores Choanocyte Amoebocyte (feeding cell) Skeletal Water fiber flow Central cavity Flagella Choanocyte in contact with an amoebocyte Sponges - Porifera • Sessile filter feeder • No mouth • Sac-like body, perforated by pores. • Interior lined by flagellated cells (choanocytes). Flagellated collar cells generate a current, draw water through the walls of the sponge where food is collected. • Amoeboid cells move around in the mesophyll and distribute food. Sponges - Porifera Grantia x.s. Sponge Reproduction Asexual reproduction • Fragmentation or by budding. • Sponges are capable of regeneration, growth of a whole from a small part. Sexual reproduction • Hermaphrodites, produce both eggs and sperm • Eggs and sperm released into the central cavity • Produces -

"Phoronida". In: Encyclopedia of Life Science
Phoronida Introductory article Christian C Emig, Centre d’Oce´anologie de Marseille CNRS, Marseille, France Article Contents . Basic Design The Phoronida, divided into two genera and 10 species, is a small marine group, which . Diversity and Lifestyles belongs to the phylum Lophophorata. Fossil History and Phylogeny Basic Design complexity of the lophophore from an oval to a helicoidal, The Phoronida is an exclusively marine group with a sessile through a horseshoe and spiral shape. This is related to an vermiform body enclosed in a tube (Figure 1) The body is increase in the number of tentacles, which is proportional composed of three distinct parts (prosome, mesosome and to the general body size. metasome), each containing its own coelomic cavity. The The metasome (or trunk) is slender and cylindrical, with prosome forms the epistome, a fold overhanging the mouth a bulb-like posterior end (or ampulla) that anchors the dorsally. The mesosome bears the lophophore, with the body in the rear end of the tube. It is separated from the rest mouth lying between its two rows of tentacles. The of the body by the diaphragm, a thick transverse septum lophophore is a terminal, bilaterally symmetrical, tentacle located behind the lophophore. crown, each tentacle having complex arrays of cilia for The digestive tract is U-shaped, bringing the anus close filter-feeding. Lophophore shape is a fairly constant to the mouth. The descending branch is divided into a short feature within each species, and there is an increasing oesophagus, followed by a long prestomach, then a stomach surrounded by a blood plexus. -

Flatworms/ Rotifers 1) Clade (Clades Are a Group of Related Phylum) Platyzoa A) Platyzoa Consist of 6 Phyla, However Most of T
Flatworms/ Rotifers 1) Clade (clades are a group of related phylum) Platyzoa a) Platyzoa consist of 6 phyla, however most of these phyla are represented by a very small number of species and are debated on where they fall taxonomically. b) These organisms represent the beginning or bilateral symmetry i) For organisms like sponges and Cnidarians it is to their advantage to be radial symmetrical because they can collect food from any angle ii) However now organisms show a distinct head and tail end and better movement to go after food items 2) Phylum Platyhelminthes a) This is the group of flat worms. b) It consist of four classes c) All but one class are parasitic in nature d) Feeding i) Platyhelminthes have an incomplete gut. ii) Most have a mouth, pharynx, and intestine (1) The advancement of having a small intestine increases surface area and thus increases the amount of nutrients absorbed. iii) Many of the non-parasitic species have a pharynx (connection between mouth and intestines) that has the ability to extend out of the mouth in order to gather resources. iv) Parasitic forms have to have some specialized feeding apparatus to extract nutrients from their host without causing too much harm. e) Sense organs i) Here is also an evolutionary advancement in nerve cells (1) The simplest forms are similar to Cnidarians, however, others have in addition one or more longitudinal nerve cords creating a “ladder” style pattern. (2) Towards the superior end there is a cluster of nerves that serves as a rudimentary brain ii) Tactile cells (cells that detect pressure) and chemoreceptors (cells that stimulate in response to a chemical) are abundant all over the body. -
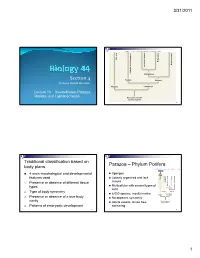
S I Section 4
3/31/2011 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Porifera Ecdysozoa Deuterostomia Lophotrochozoa Cnidaria and Ctenophora Cnidaria and Protostomia SSiection 4 Radiata Bilateria Professor Donald McFarlane Parazoa Eumetazoa Lecture 13 Invertebrates: Parazoa, Radiata, and Lophotrochozoa Ancestral colonial choanoflagellate 2 Traditional classification based on Parazoa – Phylum Porifera body plans Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Parazoa 4 main morphological and developmental Sponges features used Loosely organized and lack Porifera Ecdysozoa Cnidaria and Ctenophora tissues euterostomia photrochozoa D 1. o Presence or absence of different tissue L Multicellular with several types of types Protostomia cells 2. Type of body symmetry Radiata Bilateria 8,000 species, mostly marine Parazoa Eumetazoa 3. Presence or absence of a true body No apparent symmetry cavity Ancestral colonial Adults sessile, larvae free- choanoflagellate 4. Patterns of embryonic development swimming 3 4 1 3/31/2011 Water drawn through pores (ostia) into spongocoel Flows out through osculum Reproduce Choanocytes line spongocoel Sexually Most hermapppgggphrodites producing eggs and sperm Trap and eat small particles and plankton Gametes are derived from amoebocytes or Mesohyl between choanocytes and choanocytes epithelial cells Asexually Amoebocytes absorb food from choanocytes, Small fragment or bud may detach and form a new digest it, and carry