Proteomic and Transcriptomic Changes in Hibernating Grizzly Bears Reveal Mechanisms That Protect Against Muscle Atrophy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

9-Azido Analogs of Three Sialic Acid Forms for Metabolic Remodeling Of
Supporting Information 9-Azido Analogs of Three Sialic Acid Forms for Metabolic Remodeling of Cell-Surface Sialoglycans Bo Cheng,†,‡ Lu Dong,†,§ Yuntao Zhu,†,‡ Rongbing Huang,†,‡ Yuting Sun,†,‖ Qiancheng You,†,‡ Qitao Song,†,§ James C. Paton, ∇ Adrienne W. Paton,∇ and Xing Chen*,†,‡,§,⊥,# †College of Chemistry and Molecular Engineering, ‡Beijing National Laboratory for Molecular Sciences, §Peking−Tsinghua Center for Life Sciences,‖Academy for Advanced Interdisciplinary Studies, ⊥Synthetic and Functional Biomolecules Center, and #Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, Peking University, Beijing 100871, China ∇Research Centre for Infectious Diseases, Department of Molecular and Biomedical Science, University of Adelaide, Adelaide SA 5005, Australia Page S1 Table of Contents: Scheme S1.……………………………………………………….........……………. S3 Figure S1……………………………………………………..………..……………. S3 Figure S2……………………………………………………..………..…………… S4 Figure S3……………………………………………………..………..…………… S4 Figure S4……………………………………………………..………..…………… S5 Figure S5……………………………………………………..………..…………… S6 Figure S6……………………………………………………..………..…………….S7 Figure S7……………………………………………………..………..…………….S8 Figure S8……………………………………………………..………..…………….S9 Experimental Procedures……………………………….…........…………....S10-S27 Table S1………………………………………………..………..…………….S28-S48 Supporting Reference……………………………………………….......………...S49 Page S2 Scheme S1. Synthesis of 9AzNeu5Gc Figure S1: a, b, c, d) Representative scatter plots (FSC vs. SSC) and histograms of flow cytometry analysis -

The Concise Guide to PHARMACOLOGY 2015/16: Transporters
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: Transporters. British Journal of Pharmacology (2015) 172, 6110–6202 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: Transporters Stephen PH Alexander1, Eamonn Kelly2, Neil Marrion2, John A Peters3, Helen E Benson4, Elena Faccenda4, Adam J Pawson4, Joanna L Sharman4, Christopher Southan4, Jamie A Davies4 and CGTP Collaborators L 1 School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2 School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK 3 Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK N 4 Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13355/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: G protein-coupled receptors, ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. The Concise Guide is published in landscape format in order to facilitate comparison of related targets. -

1 Red Blood Cell Tension Protects Against Severe Malaria in The
Red blood cell tension protects against severe malaria in the Dantu blood group Silvia N. Kariuki1*, Alejandro Marin-Menendez2*, Viola Introini3*, Benjamin J. Ravenhill4, Yen- Chun Lin3, Alex Macharia1, Johnstone Makale1, Metrine Tendwa1, Wilfred Nyamu1, Jurij Kotar3, Manuela Carrasquilla2, J. Alexandra Rowe5, Kirk Rockett6, Dominic Kwiatkowski2,6,7, Michael P. Weekes4, Pietro Cicuta3$#, Thomas N. Williams1,8$#, Julian C. Rayner2,4$# 1KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya 2Wellcome Sanger Institute, Cambridge, UK 3Cavendish Laboratory, University of Cambridge, Cambridge, UK 4Cambridge Institute for Medical Research, University of Cambridge, Cambridge, UK 5University of Edinburgh, Edinburgh, UK 6Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK 7Big Data Institute, University of Oxford, Oxford, UK 8Imperial College London, London, UK * equal contributions $ jointly supervising # corresponding Pietro Cicuta, Cavendish Laboratory, University of Cambridge, Cambridge CB3 0HE, United Kingdom. Phone: +44 1223 337462. E-mail: [email protected] Thomas N. Williams, KEMRI-Wellcome Trust Research Programme, Centre for Geographic Medicine Research-Coast, Kilifi 80108, Kenya. E-mail: [email protected] 1 Julian C. Rayner, Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, United Kingdom. Phone: +44 1223 492327. E-mail: [email protected] 2 Malaria has had a major effect on the human genome, many protective polymorphisms such as sickle cell trait having been selected to high frequencies in malaria endemic regions1,2. Recently, it was shown that the blood group variant Dantu provides 74% protection against all forms of severe malaria in homozygous individuals3-5. This is a similar degree of protection to sickle cell trait and considerably greater than the best malaria vaccine, but until now the protective mechanism has been unknown. -
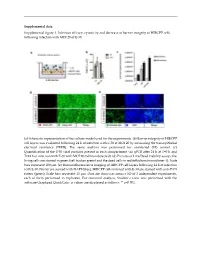
Supplemental Figure 1. Infection Efficacy, Cytoxicity and Decrease in Barrier Integrity of HIBCPP Cells Following Infection with MOI 20 of E-30
Supplemental data: Supplemental figure 1. Infection efficacy, cytoxicity and decrease in barrier integrity of HIBCPP cells following infection with MOI 20 of E-30. (a) Schematic representation of the culture model used for the experiments. (b) Barrier integrity of HIBCPP cell layers was evaluated following 24 h of infection with E-30 at MOI 20 by measuring the transepithelial electrical resistance (TEER). The same analysis was performed for uninfected (UI) control. (c) Quantification of the E-30 viral particles present in each compartment via qPCR after 24 h at T=0 h and T=24 h of infection with E-30 with MOI 20 (nd=non detected). (d) Pictures of Live/Dead viability assays; the living cells are stained in green (cell tracker green) and the dead cells in red (ethidium homodimer-1). Scale bars represent 100 µm. (e) Immunofluorescence imaging of HIBCPP cell layers following 24 h of infection with E-30. Nuclei are stained with DAPI (blue), HIBCPP cells infected with E-30 are stained with anti-PAN entero (green). Scale bars represent 15 µm. Data are shown as mean ± SD of 3 independent experiments, each of them performed in triplicates. For statistical analysis, Student’s t-test was performed with the software Graphpad QuickCalcs. p values are displayed as follows: ** p<0.001. Supplemental table 1. Up-regulated proteins in the lipid raft of HIBCPP cells following 24 h of E-30 infection at MOI 20. Proteins were identified as significantly changed in abundancy, when they exceeded a t-test difference > 1.0. Results shown are the comparison of 3 different independent proteomic experiments. -

Transporters
University of Dundee The Concise Guide to PHARMACOLOGY 2015/16 Alexander, Stephen P. H.; Kelly, Eamonn; Marrion, Neil; Peters, John A.; Benson, Helen E.; Faccenda, Elena Published in: British Journal of Pharmacology DOI: 10.1111/bph.13355 Publication date: 2015 Licence: CC BY Document Version Publisher's PDF, also known as Version of record Link to publication in Discovery Research Portal Citation for published version (APA): Alexander, S. P. H., Kelly, E., Marrion, N., Peters, J. A., Benson, H. E., Faccenda, E., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2015). The Concise Guide to PHARMACOLOGY 2015/16: Transporters. British Journal of Pharmacology, 172(24), 6110-6202. https://doi.org/10.1111/bph.13355 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 06. Oct. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: Transporters. -

New Nucleotide Sequence Data on the EMBL File Server
kQD/ 1992 Oxford University Press Nucleic Acids Research, Vol. 20, No. 13 3531-3546 New nucleotide sequence data on the EMBL File Server April 21 to May 5, 1992 New nucleotide sequence data, available from the EMBL File Server, (see Stoehr, P.J. and Omond, R.A. (1989) Nucleic Acids Res., 17 (16), 6763-6764), are reported below. Availability of all the newest sequence data is the result of collaboration between the EMBL Data Library, GenBank® and DDBJ, and data are supplied regularly by these groups. Updates to existing data are not reported here. This report has been prepared by the EMBL Data Library. The EMBL File Server can be used by anyone who has access to electronic mail networks. Database entries can be retrieved by sending a mail message to the Internet address [email protected] containing the GET command. For example, to get the entry with the accession number X12399, the message should include the following line: GET NUC:X12399 The requested file(s) will be returned automatically via electronic mail. More introductory documentation can be obtained by using the file server command: HELP PRIMATES: H.sapiens apolipoprotein A-I (APOAl) gene, exon 1 and 5' end. Ichinose A.; Homo sapiens activator 1 (replication factor C) 40 kDa subunit Biochemistry 31:3113-3118(1992). M86877 (Al) mRNA, complete cds. Chen M., Pan Z.Q., Hurwitz J.; H.sapiens apolipoprotein A-II (APOA2) gene, exon 1 and 5' end. Proc. Natl. Acad. Sci. U.S.A. 89:2516-2520(1992). M87338 Ichinose A.; Biochemistry 31:3113-3118(1992). -

UGT) 2B7 in Human Liver Microsomes
Drug Metab. Pharmacokinet. 29 (3): 259–265 (2014). Copyright © 2014 by the Japanese Society for the Study of Xenobiotics (JSSX) Regular Article Extensive Protein-Protein Interactions Involving UDP- glucuronosyltransferase (UGT) 2B7 in Human Liver Microsomes Ryoichi FUJIWARA* and Tomoo ITOH School of Pharmacy, Kitasato University, Tokyo, Japan Full text of this paper is available at http://www.jstage.jst.go.jp/browse/dmpk Summary: UDP-glucuronosyltransferase (UGT) 2B7 is a membrane protein that catalyzes glucuronidation of endogenous and exogenous substrates. Because UGTs are expressed in the endoplasmic reticulum (ER), their substrates and metabolites need to be transported through the ER membrane. However, insight into the mechanism underlying the transport of substrates/metabolites of UGTs through the ER membrane has not been elucidated. Metabolosome is a functional unit of metabolism consisting of multiple metabolism- related proteins. UGTs might form a metabolosome to facilitate the transport of their substrates and/or metabolites through the ER membrane. In the present study, therefore, extensive protein-protein inter- actions involving UGT2B7 were determined by a shotgun analysis of immunoprecipitate. Our shotgun analysis revealed that 92 proteins were immunoprecipitated with anti-UGT2B7 antibody in human liver microsomes. We further determined that 42 proteins out of the 92 proteins were specifically immuno- precipitated with the anti-UGT2B7 antibody. In addition to UGT2B7, other microsomal enzymes such as UGT1A, CYP3A4, CYP1A2, and a monoamine oxidase, were included in the list of proteins immuno- precipitated with the anti-UGT2B7 antibody, suggesting that these proteins might form a metabolosome to regulate their functions in the liver. Further analyses are needed to elucidate the roles of those proteins in the enzymatic activity of human UGTs. -

Transcriptional Profiling of a Mouse Model of Infectious Colitis: the Role of Host Factors
Development, Characterization and Transcriptional Profiling of a Mouse Model of Fatal Infectious Diarrhea and Colitis. by Diana Borenshtein M.Sc. Nutritional Sciences, 1999 B.Sc. Nutritional Sciences, 1995 The Hebrew University of Jerusalem, Israel SUBMITTED TO THE DEPARTMENT OF BIOLOGICAL ENGINEERING IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN MOLECULAR AND SYSTEMS TOXICOLOGY at the MASSACHUSETTS INSTITUTE OF TECHNOLOGY February 2007 © 2007 Massachusetts Institute of Technology. All rights reserved Signature of Author Biological Engineering Division December 1, 2006 Certified by David B. Schauer Professor of Biological Engineering Thesis Supervisor Accepted by Ram Sasisekharan Professor of Biological Engineering Chairman, Committee for Graduate Students 2 This doctoral thesis has been examined by a committee of the department of Biological Engineering as follows: Certified by Professor James G. Fox, MIT Chairman Certified by Professor James L. Sherley, MIT Certified by Professor Eric J. Rubin, Harvard School of Public Health 3 4 Development, Characterization and Transcriptional Profiling of a Mouse Model of Fatal Infectious Diarrhea and Colitis by Diana Borenshtein Submitted to the Biological Engineering Division on December 1, 2006 in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Molecular and Systems Toxicology Abstract Citrobacter rodentium is a naturally occurring murine bacterial pathogen which is used to model human diarrheagenic E. coli (EPEC and EHEC) infections in mice. C. rodentium causes colonic hyperplasia and a variable degree of colitis and mortality in the majority of inbred and outbred lines of mice. Differences in C. rodentium-induced disease are determined by the genetic background of the host. -

Protein List (6009)
Protein Accession Protein Id Protein Name Q9CQV8 1433B 14-3-3 protein beta/alpha OS=Mus musculus OX=10090 GN=Ywhab PE=1 SV=3 P62259 1433E 14-3-3 protein epsilon OS=Mus musculus OX=10090 GN=Ywhae PE=1 SV=1 P68510 1433F 14-3-3 protein eta OS=Mus musculus OX=10090 GN=Ywhah PE=1 SV=2 P61982 1433G 14-3-3 protein gamma OS=Mus musculus OX=10090 GN=Ywhag PE=1 SV=2 P68254 1433T 14-3-3 protein theta OS=Mus musculus OX=10090 GN=Ywhaq PE=1 SV=1 P63101 1433Z 14-3-3 protein zeta/delta OS=Mus musculus OX=10090 GN=Ywhaz PE=1 SV=1 Q6PD03 2A5A Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform OS=Mus musculus OX=10090 GN=Ppp2r5a PE=1 SV=1 Q6PD28 2A5B Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit beta isoform OS=Mus musculus OX=10090 GN=Ppp2r5b PE=1 SV=1 Q61151 2A5E Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform OS=Mus musculus OX=10090 GN=Ppp2r5e PE=1 SV=3 Q60996 2A5G Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform OS=Mus musculus OX=10090 GN=Ppp2r5c PE=1 SV=2 Q76MZ3 2AAA Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform OS=Mus musculus OX=10090 GN=Ppp2r1a PE=1 SV=3 Q7TNP2 2AAB Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform OS=Mus musculus OX=10090 GN=Ppp2r1b PE=1 SV=2 Q6P1F6 2ABA Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform OS=Mus musculus OX=10090 GN=Ppp2r2a PE=1 SV=1 Q925E7 2ABD Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B delta isoform OS=Mus musculus OX=10090 GN=Ppp2r2d PE=1 SV=1 P55194 3BP1 SH3 domain-binding protein 1 OS=Mus musculus OX=10090 GN=Sh3bp1 PE=1 SV=3 Q06649 3BP2 SH3 domain-binding protein 2 OS=Mus musculus OX=10090 GN=Sh3bp2 PE=1 SV=1 Q99L13 3HIDH 3-hydroxyisobutyrate dehydrogenase, mitochondrial OS=Mus musculus OX=10090 GN=Hibadh PE=1 SV=1 P48193 41 Protein 4. -

Marrion, Neil V
University of Dundee THE CONCISE GUIDE TO PHARMACOLOGY 2017/18 Alexander, Stephen P. H.; Kelly, Eamonn; Marrion, Neil V.; Peters, John A.; Faccenda, Elena; Harding, Simon D.; Pawson, Adam J.; Sharman, Joanna L.; Southan, Christopher; Davies, Jamie A.; CGTP Collaborators Published in: British Journal of Pharmacology DOI: 10.1111/bph.13883 Publication date: 2017 Document Version Publisher's PDF, also known as Version of record Link to publication in Discovery Research Portal Citation for published version (APA): Alexander, S. P. H., Kelly, E., Marrion, N. V., Peters, J. A., Faccenda, E., Harding, S. D., ... CGTP Collaborators (2017). THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: Transporters. British Journal of Pharmacology, 174 (Suppl. 1), S360-S446. DOI: 10.1111/bph.13883 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2017/18: Transporters. -

5779 Protein Identified, 4414 with 2 Or More Peptides, FDR 0.01
Proteomic anlysis of human PHEO a PGL (2 x PHEO, 1 x PGL and control adrenal medulla, iTRAQ labels 114-117 5779 protein identified, 4414 with 2 or more peptides, FDR 0.01 Coverage # Unique Accession Description [%] Peptides Q9C0C9 (E3-independent) E2 ubiquitin-conjugating enzyme OS=Homo sapiens GN=UBE2O PE=1 SV=3 7 7 O14874 [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase, mitochondrial OS=Homo sapiens GN=BCKDK PE=1 SV=2 7 2 Q8TDZ2 [F-actin]-monooxygenase MICAL1 OS=Homo sapiens GN=MICAL1 PE=1 SV=2 11 5 Q7RTP6 [F-actin]-monooxygenase MICAL3 OS=Homo sapiens GN=MICAL3 PE=1 SV=2 1 1 Q15118 [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 1, mitochondrial OS=Homo sapiens GN=PDK1 PE=1 SV=1 17 4 Q15119 [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial OS=Homo sapiens GN=PDK2 PE=1 SV=2 16 3 Q15120 [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 3, mitochondrial OS=Homo sapiens GN=PDK3 PE=1 SV=1 13 3 Q9P0J1 [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, mitochondrial OS=Homo sapiens GN=PDP1 PE=1 SV=3 29 10 Q04446 1,4-alpha-glucan-branching enzyme OS=Homo sapiens GN=GBE1 PE=1 SV=3 38 19 P61604 10 kDa heat shock protein, mitochondrial OS=Homo sapiens GN=HSPE1 PE=1 SV=2 77 10 Q15029 116 kDa U5 small nuclear ribonucleoprotein component OS=Homo sapiens GN=EFTUD2 PE=1 SV=1 32 23 Q9NRX4 14 kDa phosphohistidine phosphatase OS=Homo sapiens GN=PHPT1 PE=1 SV=1 38 3 P31946 14-3-3 protein beta/alpha OS=Homo sapiens GN=YWHAB PE=1 SV=3 65 7 P62258 14-3-3 protein epsilon OS=Homo -
Appendix C Database Derived Peptides Probed for Riiα Binding
Appendix C Database derived peptides probed for RIIα binding Table C.1: All database derived peptides probed for RIIα binding. Depicted are the block number and position of the peptide (Bl., Pos.) corresponding to Fig. C.1, the peptide sequence, its name or swissprot identifier (SP-ID) and a description (mostly abbreviated, indicated by ···) provided by the database. A + indicates RIIα-binding. Bl. Pos. Sequence Name Description 1 A01 + QIEYLAKQIVDNAIQQA AKAPis 1 A02 QIEYLAKQIPDNAIQQA AKAPis-p 1 A03 + KGADLIEEAASRIVDAVIEQVKAAG Ht31 1 A04 KGADLIEEAASRIPDAPIEQVKAAG Ht31-PP 1 A05 + PEDAELVRLSKRLVENAVLKAVQQY AKAP7δ-wt 1 A06 + PEDAELVRTSKRLVENAVLKAVQQY AKAP7δ-L304T 1 A07 PEDAELVRLSKRDVENAVLKAVQQY AKAP7δ-L308D row A 1 A08 + PEDAELVRLSKRLVENAVEKAVQQY AKAP7δ-L314E (control 1 A09 + PEDAELVRLSKRLPENAVLKAVQQY AKAP7δ-p peptides) 1 A10 PEDAELVRLSKRLPENAPLKAVQQY AKAP7δ-pp identical 1 A11 + PEDAELVRLDKRLVENAVLKAVQQY AKAP7δ-S305D for all 1 A12 + PEDAELVRLAKRLVENAVLKAVQQY AKAP7δ-S305A blocks 1 A13 + PEAAELVRLSKRLVENAVEKAVQQY AKAP7δ-L314E/D298A 1 A14 + PEDAALVRLSKRLVENAVEKAVQQY AKAP7δ-L314E/E300A 1 A15 + PEDAELVRLAKRLVENAVEKAVQQY AKAP7δ-L314E/E300A 1 A16 + PAAAELVRLAKRLVENAVEKAVQQY AKAP7δ-L314E/E297A/ D298A/S305A 1 A17 + PEDAELVRLSKRLVENAVEKAVQAY AKAP7δ-L314E/Q319A 124 Bl. Pos. Sequence SP-ID Description 1 B01 KEAAENSLVAYKAASDIAMTELPPT 143E 14-3-3 protein epsilon (14-3-3E) (Mito··· 1 B02 LSSQPTIPIVGIIAGLVLLGAVITG 1A36 HLA class I histocompatibility antigen··· 1 B03 PSSQPTVHIVGIIAGLVLLGAVITG 1A23 HLA class I histocompatibility antigen··· 1 B04 PSSQPTVPIVGIIAGLVLLGAVITG