Welcome to the 11Th Southeast Enzyme Conference
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MINUTES of the MEETING of the BOARD of REGENTS of the UNIVERSITY SYSTEM of GEORGIA College of Coastal Georgia, Brunswick, Georgia April 18-19, 2017
MINUTES OF THE MEETING OF THE BOARD OF REGENTS OF THE UNIVERSITY SYSTEM OF GEORGIA College of Coastal Georgia, Brunswick, Georgia April 18-19, 2017 CALL TO ORDER The Board of Regents of the University System of Georgia met at 10:05 a.m. Tuesday, April 18, and 9:01 a.m. Wednesday, April 19, 2017, at the College of Coastal Georgia, Brunswick, Georgia. The Chair of the Board, Regent C. Thomas Hopkins, Jr., called the meeting to order both days. Present, in addition to Chair Hopkins, were Vice Chair James M. Hull; and Regents C. Dean Alford; W. Paul Bowers; Rutledge A. Griffin, Jr.; Donald M. Leebern, Jr.; Laura Marsh; Doreen Stiles Poitevint; Neil L. Pruitt, Jr.; Sarah-Elizabeth Reed; E. Scott Smith; Richard L. Tucker; T. Rogers Wade; Larry Walker; Don L. Waters; and Philip A. Wilheit, Sr. Regents Kessel D. Stelling, Jr.; Sachin Shailendra; and Benjamin J. Tarbutton, III, were excused both days. Regents W. Paul Bowers was excused Wednesday, April 19, 2017. INVOCATION AND PLEDGE College of Coastal Georgia Student Government Association President Foster Hayes gave the invocation and led the Pledge of Allegiance both days. SAFETY BRIEFING College of Coastal Georgia Chief of Police Bryan Snipe gave the safety briefing both days. APPROVAL OF MINUTES With motion made and variously seconded, the Regents who were present voted unanimously to approve the minutes of the Board’s March 15, 2017, meeting. PRESIDENT’S PRESENTATION At approximately 10:11 a.m. Tuesday, April 18, 2017, College of Coastal Georgia President Gregory Aloia welcomed the Board of Regents and guests to the institution, and gave a presentation showcasing the school’s programs, students, and culture. -

Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications
International Journal of Molecular Sciences Review Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications Daniel Fernández-Villa 1, Maria Rosa Aguilar 1,2 and Luis Rojo 1,2,* 1 Instituto de Ciencia y Tecnología de Polímeros, Consejo Superior de Investigaciones Científicas, CSIC, 28006 Madrid, Spain; [email protected] (D.F.-V.); [email protected] (M.R.A.) 2 Consorcio Centro de Investigación Biomédica en Red de Bioingeniería, Biomateriales y Nanomedicina, 28029 Madrid, Spain * Correspondence: [email protected]; Tel.: +34-915-622-900 Received: 18 September 2019; Accepted: 7 October 2019; Published: 9 October 2019 Abstract: Bacterial, protozoan and other microbial infections share an accelerated metabolic rate. In order to ensure a proper functioning of cell replication and proteins and nucleic acids synthesis processes, folate metabolism rate is also increased in these cases. For this reason, folic acid antagonists have been used since their discovery to treat different kinds of microbial infections, taking advantage of this metabolic difference when compared with human cells. However, resistances to these compounds have emerged since then and only combined therapies are currently used in clinic. In addition, some of these compounds have been found to have an immunomodulatory behavior that allows clinicians using them as anti-inflammatory or immunosuppressive drugs. Therefore, the aim of this review is to provide an updated state-of-the-art on the use of antifolates as antibacterial and immunomodulating agents in the clinical setting, as well as to present their action mechanisms and currently investigated biomedical applications. Keywords: folic acid antagonists; antifolates; antibiotics; antibacterials; immunomodulation; sulfonamides; antimalarial 1. -

Reduction by the Methylreductase System in Methanobacterium Bryantii WILLIAM B
JOURNAL OF BACTERIOLOGY, Jan. 1987, p. 87-92 Vol. 169, No. 1 0021-9193/87/010087-06$02.00/0 Copyright © 1987, American Society for Microbiology Inhibition by Corrins of the ATP-Dependent Activation and CO2 Reduction by the Methylreductase System in Methanobacterium bryantii WILLIAM B. WHITMAN'* AND RALPH S. WOLFE2 Department of Microbiology, University of Georgia, Athens, Georgia 30602,1 and Department of Microbiology, University ofIllinois, Urbana, Illinois 618012 Received 1 August 1986/Accepted 28 September 1986 Corrins inhibited the ATP-dependent activation of the methylreductase system and the methyl coenzyme M-dependent reduction of CO2 in extracts of Methanobacterium bryantii resolved from low-molecular-weight factors. The concentrations of cobinamides and cobamides required for one-half of maximal inhibition of the ATP-depen4ent activation were between 1 and 5 ,M. Cobinamides were more inhibitory at lower concentra- tiops than cobamides. Deoxyadenosylcobalamin was not inhibitory at concentrations up to 25 ,uM. The inhibition of CO2 reduction was competitive with respect to CO2. The concentration of methylcobalamin required for one-half of maximal inhibition was 5 ,M. Other cobamideg inhibited at similar concentrations, but diaquacobinami4e inhibited at lower concentrations. With respect to their affinities and specificities for corrins, inhibition of both the ATP-dependent activation'and CO2 reduction closely resembled the corrin- dependent activation of the methylreductase described in similar extracts (W. B. Whitman and R. S. Wolfe, J. Bacteriol. 164:165-172, 1985). However, whether the multiple effects of corrins are due to action at a single site is unknown. The effect of corrins (cobamides and cobinamides) on in CO2 reduction. -

Kelechi Nmaobi Uzochukwu
Kelechi N. Uzochukwu, PhD Assistant Professor www.kelechiuzo.webs.com | [email protected] | (410) 837-5061 U.S. Citizen EDUCATION 2014 PhD, Public Policy Atlanta, GA Georgia State University & Georgia Institute of Technology (Joint PhD Program) Specializations: Planning & Economic Development | Public & Nonprofit Management Dissertation: “Assessing the Prevalence, Participants, and Predictors of Coproduction: The Case of Atlanta, Georgia” – Advisor John Clayton Thomas 2007 MPA, Master of Public Administration Atlanta, GA Andrew Young School of Policy Studies, Georgia State University Specialization: Management & Finance 2004 BS, Civil & Environmental Engineering Greensboro, NC North Carolina Agricultural & Technical State University Waste Management Certification RESEARCH & TEACHING INTERESTS . Urban/Community Planning . Politics & Policies . GIS Applications . Research Methods . Race/Gender/Class Issues . Public Participation . Policy Analysis . Program Evaluation PUBLICATIONS Uzochukwu, K. 2015. “Citizen Engagement in Community Development.” Federal Reserve Bank of Atlanta, Partners Update (November/December 2015). Uzochukwu, K. 2014. “The Associations between Neighborhood Constructs and Physical Activity: Understanding Race & Income Disparities.” (revise & resubmit at Journal of Urban Affairs) 2014 Best Research Paper Award, Ivan Allen College of Liberal Arts, Georgia Tech Uzochukwu, K. 2013. “Conjunction Junction, What’s Your Function? An Assessment of Coproduction in Atlanta, Georgia.” Public Administration Times. Hubsmith, D. and K. Uzochukwu. 2013. “Safe Routes to Schools.” In Encyclopedia of School Health (Vol. 1), eds. D.C. Wiley and A.C. Cory. Sage Publications, pp. 517-519. In Preparation Uzochukwu, K., Thomas, J.C. “Who Engages in the Co-production of Public Services and Why? The Case of Atlanta, Georgia” Uzochukwu, K. “Comparing the Effectiveness of Formal and Informal Coproduction in Engaging Underrepresented Groups in Public Service Delivery” Uzochukwu, K. -

Georgia State University Complete College Georgia
2016 Status Report Georgia State University Complete College Georgia Overview When it comes to higher education, the vision of the United States as a land of equal opportunity is far from a reality. Today, it is eight times more likely that an individual in the top quartile of Americans by annual household income will hold a college degree than an individual in the lowest quartile.1 Nationally, white students graduate from college at rates more than 10 points higher than Hispanic students, and are more than twice as likely to graduate with a 4-year college degree compared to black students.2 The United States Department of Education cites a six-year graduation-rate of 39% among Pell-eligible students,3 a rate that is 20 points lower than the national average.4 In 2003, Georgia State’s institutional graduation rate stood at 32% and underserved populations were foundering. Graduation rates were 22% for Latinos, 29% for African Americans, and 18% for African American males. Pell students were graduating at rates far below those of non-Pell students. Today, thanks to a campus-wide commitment to student success and more than a dozen strategic programs implemented over the past several years, Georgia State’s achievement gap is gone. The graduation rate for bachelor-degree seeking students has improved 22 points—among the highest increases in the nation over this period (Chart 1). (See Appendix for all charts.) Rates are up 36 points for Latinos (to 58%), and 29 points for African Americans (to 58%). Pell-eligible students currently represent 58% of Georgia State University’s undergraduate student population, and over the past three years have graduated at rates, on average, equal to those of non-Pell students. -
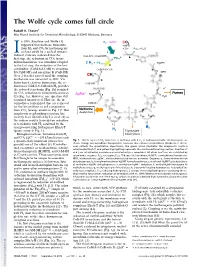
The Wolfe Cycle Comes Full Circle
The Wolfe cycle comes full circle Rudolf K. Thauer1 Max Planck Institute for Terrestrial Microbiology, D-35043 Marburg, Germany n 1988, Rouvière and Wolfe (1) H - ΔμNa+ 2 CO2 suggested that methane formation + MFR from H and CO by methanogenic + 2H+ *Fd + H O I 2 2 ox 2 archaea could be a cyclical process. j O = Indirect evidence indicated that the CoB-SH + CoM-SH fi *Fd 2- a rst step, the reduction of CO2 to for- red R mylmethanofuran, was somehow coupled + * H MPT 2 H2 Fdox 4 to the last step, the reduction of the het- h erodisulfide (CoM-S-S-CoB) to coenzyme CoM-S-S-CoB b MFR M (CoM-SH) and coenzyme B (CoB-SH). H Over 2 decades passed until the coupling C 4 10 mechanism was unraveled in 2011: Via g flavin-based electron bifurcation, the re- CoB-SH duction of CoM-S-S-CoB with H provides 2 H+ the reduced ferredoxin (Fig. 1h) required c + Purines for CO2 reduction to formylmethanofuran ΔμNa + H MPT 4 f H O (2) (Fig. 1a). However, one question still 2 remained unanswered: How are the in- termediates replenished that are removed CoM-SH for the biosynthesis of cell components H Methionine d from CO2 (orange arrows in Fig. 1)? This Acetyl-CoA e anaplerotic (replenishing) reaction has F420 F420H2 recently been identified by Lie et al. (3) as F420 F420H2 the sodium motive force-driven reduction H i of ferredoxin with H2 catalyzed by the i energy-converting hydrogenase EhaA-T H2 (green arrow in Fig. -

Identification and Characterization of Andalusicin: N-Terminally Dimethylated Class III Lantibiotic from Bacillus Thuringiensis Sv
Identification and characterization of andalusicin: N-terminally dimethylated class III lantibiotic from Bacillus thuringiensis sv. andalousiensis Anastasiia Grigoreva, Julia Andreeva, Dmitry Bikmetov, Anastasiia Rusanova, Marina Serebryakova, Andrea Hernandez Garcia, Darya Slonova, Satish Nair, Guy Lippens, Konstantin Severinov, et al. To cite this version: Anastasiia Grigoreva, Julia Andreeva, Dmitry Bikmetov, Anastasiia Rusanova, Marina Serebryakova, et al.. Identification and characterization of andalusicin: N-terminally dimethylated class III lantibiotic from Bacillus thuringiensis sv. andalousiensis. iScience, Elsevier, 2021, 24 (5), 10.1016/j.isci.2021.102480. hal-03270626 HAL Id: hal-03270626 https://hal.inrae.fr/hal-03270626 Submitted on 25 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License iScience ll OPEN ACCESS Article Identification and characterization of andalusicin: N-terminally dimethylated class III lantibiotic from Bacillus thuringiensis sv. andalousiensis -

1 SUPPLEMENTARY DATA Table S1
Electronic Supplementary Material (ESI) for Food & Function. This journal is © The Royal Society of Chemistry 2020 SUPPLEMENTARY DATA Table S1: Effects of catalase, protease, papain and pH treatments on the antimicrobial activity (%) of the CFS of S. thermophilus strain against G. vaginalis ATCC 14018. The antimicrobial activity of the strain was highest under acidic pH conditions. Remaining antimicrobial activity (%) Strains 5 mg mL-1 1 mg mL-1 1 mg mL-1 pH treatments catalase proteinase K papain 3.5 4.0 5.0 6.0 6.5 7.0 S. thermophilus KLDS 3.1003 90.49±0.62 100±0 100±0 100±0 100±0 67.02±0.08 6.35±0.03 0±0 0±0 All values were determined in triplicate Values were expressed as mean ± SD 1 Table S2: Weekly weights of study animals before and after S. aureus ATCC25923 infection. Significant variations (at P > 0.05) were observed in the weight of study animals during the study. Period C C C C Week 0 22.00b 23.57b 19.34e 23.24a 20.90d 22.84c 20.19d 22.60b 23.92a 23.70b 20.56c 23.78a 20.89d 23.10c 20.00d 23.51a Average 22.43 23.30 20.02 23.28 SD 1.29 0.40 0.51 0.51 C TST TSA TSTSA 24.92a 24.15a 21.19c 20.98c Week 1 23.34a 23.13c 20.26d 22.68b 21.98b 22.96d 20.85c 22.86b 22.60b 22.36d 24.28a 22.33b Average 23.21 23.15 21.65 22.21 SD 1.27 0.74 1.80 0.86 23.25a 24.28a 19.94d 20.39d Week 2 21.32c 22.09e 18.46f 20.35d 20.29d 21.82e 19.40e 16.68e 21.27c 22.89d 22.74b 20.09d Average 21.53 22.77 20.14 19.38 SD 1.24 1.10 1.84 1.80 Values with the same alphabet along the same column are not significantly different (P > 0.05) Composition of control and trial diets are given in Table 1. -

MITOCHONDRIAL CREATINE KINASE Some Clinical, Biochemical and Morphological Aspects
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/114105 Please be advised that this information was generated on 2021-10-11 and may be subject to change. MITOCHONDRIAL CREATINE KINASE some clinical, biochemical and morphological aspects Jan A.M. Smeitink MITOCHONDRIAL CREATINE KINASE some clinical, biochemical and morphological aspects Jan A.M. Smeitink MITOCHONDRIAL CREATINE KINASE SOME CLINICAL, BIOCHEMICAL AND MORPHOLOGICAL ASPECTS EEN WETENSCHAPPELIJKE PROEVE OP HET GEBIED VAN DE MEDISCHE WETENSCHAPPEN, IN HET BIJZONDER DE GENEESKUNDE PROEFSCHRIFT TER VERKRIJGING VAN DE GRAAD VAN DOCTOR AAN DE KATHOLIEKE UNIVERSITEIT NIJMEGEN VOLGENS BESLUIT VAN HET COLLEGE VAN DECANEN IN HET OPENBAAR TE VERDEDIGEN OP DINSDAG 6 OKTOBER 1992, DES NAMIDDAGS TE 1.30 UUR PRECIES DOOR JOHANNES ALBERTUS MARIA SMEITINK GEBOREN OP 21 JUNI 1956 TE ARNHEM IV Promotores : Prof. Dr. R.C.A. Sengers Prof. Dr. J.M.F. Trijbels Co-Promotores : Dr. W. Ruitenbeek Dr. R.A. Wevers Aan mijn ouders AanWillemien en Mark CONTEN CHAPTER 1 Introduction and aim of the study CHAPTER 2 Mitochondrial creatine kinase: a key enzyme of aerobic energy metabolism Biochimica et Biophysica Acta (Reviews on Bioenergetics): in press I. Introduction II. Biochemical studies of Mi-CK ΠΙ. Functional studies of Mi-CK IV. Integration of Mi-CK in cellular energy metabolism V. Perspectives CHAPTER 3 A method for quantitative measurement -

Lombricine Kinase Structure and Substrate Specificity: a Paradigm for Elucidation of Substrate Specificity in Phosphagen Kinases D
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2007 Lombricine Kinase Structure and Substrate Specificity: A Paradigm for Elucidation of Substrate Specificity in Phosphagen Kinases D. Jeffrey. Bush Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] THE FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES LOMBRICINE KINASE STRUCTURE AND SUBSTRATE SPECIFICITY: A PARADIGM FOR ELUCIDATION OF SUBSTRATE SPECIFICITY IN PHOSPHAGEN KINASES By D. JEFFREY BUSH A Dissertation submitted to the Department of Chemistry and Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Spring Semester, 2007 The members of the Committee approve the Dissertation of D. Jeffrey Bush defended on February 20, 2007. Michael S. Chapman Professor Co-Directing Dissertation John Dorsey Professor Co-Directing Dissertation W. Ross Ellington Outside Committee Member Michael Blaber Committee Member Approved: ____________________________________________ Joseph Schlenoff, Department Chair, Department of Chemistry & Biochemistry ____________________________________________ Joseph Travis, Dean, College of Arts & Sciences The Office of Graduate Studies has verified and approved the above named committee members. ii To the late Clifford M. Bush, who with statements such as “A heterogeneous compound of two or more substances whose ray through certain limits is confined to a specific area…” fostered a strong interest of the author in science at a very young age, if only just to know more about what he spoke. iii ACKNOWLEDGEMENTS I wish to first convey my sincere gratitude to my parents, Donald and Roberta for raising me in the nurture and admonition of the Almighty God. -

Acidophilic Green Algal Genome Provides Insights Into Adaptation to an Acidic Environment
Acidophilic green algal genome provides insights into adaptation to an acidic environment Shunsuke Hirookaa,b,1, Yuu Hirosec, Yu Kanesakib,d, Sumio Higuchie, Takayuki Fujiwaraa,b,f, Ryo Onumaa, Atsuko Eraa,b, Ryudo Ohbayashia, Akihiro Uzukaa,f, Hisayoshi Nozakig, Hirofumi Yoshikawab,h, and Shin-ya Miyagishimaa,b,f,1 aDepartment of Cell Genetics, National Institute of Genetics, Shizuoka 411-8540, Japan; bCore Research for Evolutional Science and Technology, Japan Science and Technology Agency, Saitama 332-0012, Japan; cDepartment of Environmental and Life Sciences, Toyohashi University of Technology, Aichi 441-8580, Japan; dNODAI Genome Research Center, Tokyo University of Agriculture, Tokyo 156-8502, Japan; eResearch Group for Aquatic Plants Restoration in Lake Nojiri, Nojiriko Museum, Nagano 389-1303, Japan; fDepartment of Genetics, Graduate University for Advanced Studies, Shizuoka 411-8540, Japan; gDepartment of Biological Sciences, Graduate School of Science, University of Tokyo, Tokyo 113-0033, Japan; and hDepartment of Bioscience, Tokyo University of Agriculture, Tokyo 156-8502, Japan Edited by Krishna K. Niyogi, Howard Hughes Medical Institute, University of California, Berkeley, CA, and approved August 16, 2017 (received for review April 28, 2017) Some microalgae are adapted to extremely acidic environments in pumps that biotransform arsenic and archaeal ATPases, which which toxic metals are present at high levels. However, little is known probably contribute to the algal heat tolerance (8). In addition, the about how acidophilic algae evolved from their respective neutrophilic reduction in the number of genes encoding voltage-gated ion ancestors by adapting to particular acidic environments. To gain channels and the expansion of chloride channel and chloride car- insights into this issue, we determined the draft genome sequence rier/channel families in the genome has probably contributed to the of the acidophilic green alga Chlamydomonas eustigma and per- algal acid tolerance (8). -

192ICM ICBIC Posters
Journal of Inorganic Biochemistry 96 (2003) 203 Monomeric TpPrMoVOSR complexes via the chemical reduction of TpPrMoVIOSR. David J Nielsen, School of Chemistry, University of Melbourne, Australia Christian J Doonan, School of Chemistry, University of Melbourne, Australia Graham N George, Stanford Synchrotron Radiation Laboratory, United States Hugh Harris, Stanford Synchrotron Radiation Laboratory, United States Charles G Young, University of Melbourne, Australia EPR evidence has suggested the presence of molybdenum(V) intermediates in the catalytic cycle of hydroxylase enzyme systems [1], and references therein], and as such these species are attractive targets for the synthesis of small-molecule model systems. Ongoing work in our group has allowed access to several stable and well characterised monomeric molybdenum(VI) oxo-thio complexes TpPrMoVIOSR (TpPr = hydridotris(3-isopropylpyrazol-1-yl)borate) with co-ligand R = eg. substituted phenolates [2], as shown below. These Mo(VI) complexes have proved amenable to chemical reduction using cobaltocene (CoCp2) yielding initially the Pr V [CoCp2][Tp Mo OSR] salts [1,2]. Solution and solid state sulfur X-ray absorption spectroscopy (XAS) on selected examples of the chemically reduced species shows pre-edge features attributable to the S 1s → Mo=S π* transition of a [MoOS]+ unit. Further spectroscopic investigations (EPR, IR) are consistent with the presence of a paramagnetic Mo(V) centre bearing a terminal thio ligand. Continuing spectroscopic, structural and reactivity investigations centred on these important species will be presented. References: [1] P. D. Smith, D. A. Slizys, G. N. George and C. G. Young, J. Amer. Chem. Soc., 122(12), 2000, 2946. [2] C. J. Doonan, Unpublished results.