A Comparison of Different R-Tree Construction Techniques for Range Queries on Neuromorphological Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Suffix Trees
This material takes about 1.5 hours. 1 Suffix Trees Gusfield: Algorithms on Strings, Trees, and Sequences. Weiner 73 “Linear Pattern-matching algorithms” IEEE conference on automata and switching theory McCreight 76 “A space-economical suffix tree construction algorithm” JACM 23(2) 1976 Chen and Seifras 85 “Efficient and Elegegant Suffix tree construction” in Apos- tolico/Galil Combninatorial Algorithms on Words Another “search” structure, dedicated to strings. Basic problem: match a “pattern” (of length m) to “text” (of length n) • goal: decide if a given string (“pattern”) is a substring of the text • possibly created by concatenating short ones, eg newspaper • application in IR, also computational bio (DNA seqs) • if pattern avilable first, can build DFA, run in time linear in text • if text available first, can build suffix tree, run in time linear in pattern. • applications in computational bio. First idea: binary tree on strings. Inefficient because run over pattern many times. • fractional cascading? • realize only need one character at each node! Tries: • used to store dictionary of strings • trees with children indexed by “alphabet” • time to search equal length of query string • insertion ditto. • optimal, since even hashing requires this time to hash. • but better, because no “hash function” computed. • space an issue: – using array increases stroage cost by |Σ| – using binary tree on alphabet increases search time by log |Σ| 1 – ok for “const alphabet” – if really fussy, could use hash-table at each node. • size in worst case: sum of word lengths (so pretty much solves “dictionary” problem. But what about substrings? • Relevance to DNA searches • idea: trie of all n2 substrings • equivalent to trie of all n suffixes. -

Tree-Combined Trie: a Compressed Data Structure for Fast IP Address Lookup
(IJACSA) International Journal of Advanced Computer Science and Applications, Vol. 6, No. 12, 2015 Tree-Combined Trie: A Compressed Data Structure for Fast IP Address Lookup Muhammad Tahir Shakil Ahmed Department of Computer Engineering, Department of Computer Engineering, Sir Syed University of Engineering and Technology, Sir Syed University of Engineering and Technology, Karachi Karachi Abstract—For meeting the requirements of the high-speed impact their forwarding capacity. In order to resolve two main Internet and satisfying the Internet users, building fast routers issues there are two possible solutions one is IPv6 IP with high-speed IP address lookup engine is inevitable. addressing scheme and second is Classless Inter-domain Regarding the unpredictable variations occurred in the Routing or CIDR. forwarding information during the time and space, the IP lookup algorithm should be able to customize itself with temporal and Finding a high-speed, memory-efficient and scalable IP spatial conditions. This paper proposes a new dynamic data address lookup method has been a great challenge especially structure for fast IP address lookup. This novel data structure is in the last decade (i.e. after introducing Classless Inter- a dynamic mixture of trees and tries which is called Tree- Domain Routing, CIDR, in 1994). In this paper, we will Combined Trie or simply TC-Trie. Binary sorted trees are more discuss only CIDR. In addition to these desirable features, advantageous than tries for representing a sparse population reconfigurability is also of great importance; true because while multibit tries have better performance than trees when a different points of this huge heterogeneous structure of population is dense. -

Balanced Trees Part One
Balanced Trees Part One Balanced Trees ● Balanced search trees are among the most useful and versatile data structures. ● Many programming languages ship with a balanced tree library. ● C++: std::map / std::set ● Java: TreeMap / TreeSet ● Many advanced data structures are layered on top of balanced trees. ● We’ll see several later in the quarter! Where We're Going ● B-Trees (Today) ● A simple type of balanced tree developed for block storage. ● Red/Black Trees (Today/Thursday) ● The canonical balanced binary search tree. ● Augmented Search Trees (Thursday) ● Adding extra information to balanced trees to supercharge the data structure. Outline for Today ● BST Review ● Refresher on basic BST concepts and runtimes. ● Overview of Red/Black Trees ● What we're building toward. ● B-Trees and 2-3-4 Trees ● Simple balanced trees, in depth. ● Intuiting Red/Black Trees ● A much better feel for red/black trees. A Quick BST Review Binary Search Trees ● A binary search tree is a binary tree with 9 the following properties: 5 13 ● Each node in the BST stores a key, and 1 6 10 14 optionally, some auxiliary information. 3 7 11 15 ● The key of every node in a BST is strictly greater than all keys 2 4 8 12 to its left and strictly smaller than all keys to its right. Binary Search Trees ● The height of a binary search tree is the 9 length of the longest path from the root to a 5 13 leaf, measured in the number of edges. 1 6 10 14 ● A tree with one node has height 0. -
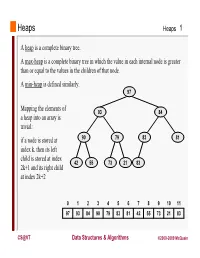
Heaps a Heap Is a Complete Binary Tree. a Max-Heap Is A
Heaps Heaps 1 A heap is a complete binary tree. A max-heap is a complete binary tree in which the value in each internal node is greater than or equal to the values in the children of that node. A min-heap is defined similarly. 97 Mapping the elements of 93 84 a heap into an array is trivial: if a node is stored at 90 79 83 81 index k, then its left child is stored at index 42 55 73 21 83 2k+1 and its right child at index 2k+2 01234567891011 97 93 84 90 79 83 81 42 55 73 21 83 CS@VT Data Structures & Algorithms ©2000-2009 McQuain Building a Heap Heaps 2 The fact that a heap is a complete binary tree allows it to be efficiently represented using a simple array. Given an array of N values, a heap containing those values can be built, in situ, by simply “sifting” each internal node down to its proper location: - start with the last 73 73 internal node * - swap the current 74 81 74 * 93 internal node with its larger child, if 79 90 93 79 90 81 necessary - then follow the swapped node down 73 * 93 - continue until all * internal nodes are 90 93 90 73 done 79 74 81 79 74 81 CS@VT Data Structures & Algorithms ©2000-2009 McQuain Heap Class Interface Heaps 3 We will consider a somewhat minimal maxheap class: public class BinaryHeap<T extends Comparable<? super T>> { private static final int DEFCAP = 10; // default array size private int size; // # elems in array private T [] elems; // array of elems public BinaryHeap() { . -

CSCI 333 Data Structures Binary Trees Binary Tree Example Full And
Notes with the dark blue background CSCI 333 were prepared by the textbook author Data Structures Clifford A. Shaffer Chapter 5 Department of Computer Science 18, 20, 23, and 25 September 2002 Virginia Tech Copyright © 2000, 2001 Binary Trees Binary Tree Example A binary tree is made up of a finite set of Notation: Node, nodes that is either empty or consists of a children, edge, node called the root together with two parent, ancestor, binary trees, called the left and right descendant, path, subtrees, which are disjoint from each depth, height, level, other and from the root. leaf node, internal node, subtree. Full and Complete Binary Trees Full Binary Tree Theorem (1) Full binary tree: Each node is either a leaf or Theorem: The number of leaves in a non-empty internal node with exactly two non-empty children. full binary tree is one more than the number of internal nodes. Complete binary tree: If the height of the tree is d, then all leaves except possibly level d are Proof (by Mathematical Induction): completely full. The bottom level has all nodes to the left side. Base case: A full binary tree with 1 internal node must have two leaf nodes. Induction Hypothesis: Assume any full binary tree T containing n-1 internal nodes has n leaves. 1 Full Binary Tree Theorem (2) Full Binary Tree Corollary Induction Step: Given tree T with n internal Theorem: The number of null pointers in a nodes, pick internal node I with two leaf children. non-empty tree is one more than the Remove I’s children, call resulting tree T’. -

Tree Structures
Tree Structures Definitions: o A tree is a connected acyclic graph. o A disconnected acyclic graph is called a forest o A tree is a connected digraph with these properties: . There is exactly one node (Root) with in-degree=0 . All other nodes have in-degree=1 . A leaf is a node with out-degree=0 . There is exactly one path from the root to any leaf o The degree of a tree is the maximum out-degree of the nodes in the tree. o If (X,Y) is a path: X is an ancestor of Y, and Y is a descendant of X. Root X Y CSci 1112 – Algorithms and Data Structures, A. Bellaachia Page 1 Level of a node: Level 0 or 1 1 or 2 2 or 3 3 or 4 Height or depth: o The depth of a node is the number of edges from the root to the node. o The root node has depth zero o The height of a node is the number of edges from the node to the deepest leaf. o The height of a tree is a height of the root. o The height of the root is the height of the tree o Leaf nodes have height zero o A tree with only a single node (hence both a root and leaf) has depth and height zero. o An empty tree (tree with no nodes) has depth and height −1. o It is the maximum level of any node in the tree. CSci 1112 – Algorithms and Data Structures, A. -

Binary Search Tree
ADT Binary Search Tree! Ellen Walker! CPSC 201 Data Structures! Hiram College! Binary Search Tree! •" Value-based storage of information! –" Data is stored in order! –" Data can be retrieved by value efficiently! •" Is a binary tree! –" Everything in left subtree is < root! –" Everything in right subtree is >root! –" Both left and right subtrees are also BST#s! Operations on BST! •" Some can be inherited from binary tree! –" Constructor (for empty tree)! –" Inorder, Preorder, and Postorder traversal! •" Some must be defined ! –" Insert item! –" Delete item! –" Retrieve item! The Node<E> Class! •" Just as for a linked list, a node consists of a data part and links to successor nodes! •" The data part is a reference to type E! •" A binary tree node must have links to both its left and right subtrees! The BinaryTree<E> Class! The BinaryTree<E> Class (continued)! Overview of a Binary Search Tree! •" Binary search tree definition! –" A set of nodes T is a binary search tree if either of the following is true! •" T is empty! •" Its root has two subtrees such that each is a binary search tree and the value in the root is greater than all values of the left subtree but less than all values in the right subtree! Overview of a Binary Search Tree (continued)! Searching a Binary Tree! Class TreeSet and Interface Search Tree! BinarySearchTree Class! BST Algorithms! •" Search! •" Insert! •" Delete! •" Print values in order! –" We already know this, it#s inorder traversal! –" That#s why it#s called “in order”! Searching the Binary Tree! •" If the tree is -

Effect of Distance Measures on Partitional Clustering Algorithms
Sesham Anand et al, / (IJCSIT) International Journal of Computer Science and Information Technologies, Vol. 6 (6) , 2015, 5308-5312 Effect of Distance measures on Partitional Clustering Algorithms using Transportation Data Sesham Anand#1, P Padmanabham*2, A Govardhan#3 #1Dept of CSE,M.V.S.R Engg College, Hyderabad, India *2Director, Bharath Group of Institutions, BIET, Hyderabad, India #3Dept of CSE, SIT, JNTU Hyderabad, India Abstract— Similarity/dissimilarity measures in clustering research of algorithms, with an emphasis on unsupervised algorithms play an important role in grouping data and methods in cluster analysis and outlier detection[1]. High finding out how well the data differ with each other. The performance is achieved by using many data index importance of clustering algorithms in transportation data has structures such as the R*-trees.ELKI is designed to be easy been illustrated in previous research. This paper compares the to extend for researchers in data mining particularly in effect of different distance/similarity measures on a partitional clustering algorithm kmedoid(PAM) using transportation clustering domain. ELKI provides a large collection of dataset. A recently developed data mining open source highly parameterizable algorithms, in order to allow easy software ELKI has been used and results illustrated. and fair evaluation and benchmarking of algorithms[1]. Data mining research usually leads to many algorithms Keywords— clustering, transportation Data, partitional for similar kind of tasks. If a comparison is to be made algorithms, cluster validity, distance measures between these algorithms.In ELKI, data mining algorithms and data management tasks are separated and allow for an I. INTRODUCTION independent evaluation. -

6.172 Lecture 19 : Cache-Oblivious B-Tree (Tokudb)
How TokuDB Fractal TreeTM Indexes Work Bradley C. Kuszmaul Guest Lecture in MIT 6.172 Performance Engineering, 18 November 2010. 6.172 —How Fractal Trees Work 1 My Background • I’m an MIT alum: MIT Degrees = 2 × S.B + S.M. + Ph.D. • I was a principal architect of the Connection Machine CM-5 super computer at Thinking Machines. • I was Assistant Professor at Yale. • I was Akamai working on network mapping and billing. • I am research faculty in the SuperTech group, working with Charles. 6.172 —How Fractal Trees Work 2 Tokutek A few years ago I started collaborating with Michael Bender and Martin Farach-Colton on how to store data on disk to achieve high performance. We started Tokutek to commercialize the research. 6.172 —How Fractal Trees Work 3 I/O is a Big Bottleneck Sensor Query Systems include Sensor Disk Query sensors and Sensor storage, and Query want to perform Millions of data elements arrive queries on per second Query recently arrived data using indexes. recent data. Sensor 6.172 —How Fractal Trees Work 4 The Data Indexing Problem • Data arrives in one order (say, sorted by the time of the observation). • Data is queried in another order (say, by URL or location). Sensor Query Sensor Disk Query Sensor Query Millions of data elements arrive per second Query recently arrived data using indexes. Sensor 6.172 —How Fractal Trees Work 5 Why Not Simply Sort? • This is what data Data Sorted by Time warehouses do. • The problem is that you Sort must wait to sort the data before querying it: Data Sorted by URL typically an overnight delay. -
B-Trees M-Ary Search Tree Solution
M-ary Search Tree B-Trees • Maximum branching factor of M • Complete tree has height = Section 4.7 in Weiss # disk accesses for find : Runtime of find : 2 Solution: B-Trees B-Trees • specialized M-ary search trees What makes them disk-friendly? • Each node has (up to) M-1 keys: 1. Many keys stored in a node – subtree between two keys x and y contains leaves with values v such that • All brought to memory/cache in one access! 3 7 12 21 x ≤ v < y 2. Internal nodes contain only keys; • Pick branching factor M Only leaf nodes contain keys and actual data such that each node • The tree structure can be loaded into memory takes one full irrespective of data object size {page, block } x<3 3≤x<7 7≤x<12 12 ≤x<21 21 ≤x • Data actually resides in disk of memory 3 4 B-Tree: Example B-Tree Properties ‡ B-Tree with M = 4 (# pointers in internal node) and L = 4 (# data items in leaf) – Data is stored at the leaves – All leaves are at the same depth and contain between 10 40 L/2 and L data items – Internal nodes store up to M-1 keys 3 15 20 30 50 – Internal nodes have between M/2 and M children – Root (special case) has between 2 and M children (or root could be a leaf) 1 2 10 11 12 20 25 26 40 42 AB xG 3 5 6 9 15 17 30 32 33 36 50 60 70 Data objects, that I’ll Note: All leaves at the same depth! ignore in slides 5 ‡These are technically B +-Trees 6 1 Example, Again B-trees vs. -

AVL Trees and Rotations
/ AVL trees and rotations Q1 Operations (insert, delete, search) are O(height) Tree height is O(log n) if perfectly balanced ◦ But maintaining perfect balance is O(n) Height-balanced trees are still O(log n) ◦ For T with height h, N(T) ≤ Fib(h+3) – 1 ◦ So H < 1.44 log (N+2) – 1.328 * AVL (Adelson-Velskii and Landis) trees maintain height-balance using rotations Are rotations O(log n)? We’ll see… / or = or \ Different representations for / = \ : Just two bits in a low-level language Enum in a higher-level language / Assume tree is height-balanced before insertion Insert as usual for a BST Move up from the newly inserted node to the lowest “unbalanced” node (if any) ◦ Use the balance code to detect unbalance - how? Do an appropriate rotation to balance the sub-tree rooted at this unbalanced node For example, a single left rotation: Two basic cases ◦ “See saw” case: Too-tall sub-tree is on the outside So tip the see saw so it’s level ◦ “Suck in your gut” case: Too-tall sub-tree is in the middle Pull its root up a level Q2-3 Unbalanced node Middle sub-tree attaches to lower node of the “see saw” Diagrams are from Data Structures by E.M. Reingold and W.J. Hansen Q4-5 Unbalanced node Pulled up Split between the nodes pushed down Weiss calls this “right-left double rotation” Q6 Write the method: static BalancedBinaryNode singleRotateLeft ( BalancedBinaryNode parent, /* A */ BalancedBinaryNode child /* B */ ) { } Returns a reference to the new root of this subtree. -

Tries and String Matching
Tries and String Matching Where We've Been ● Fundamental Data Structures ● Red/black trees, B-trees, RMQ, etc. ● Isometries ● Red/black trees ≡ 2-3-4 trees, binomial heaps ≡ binary numbers, etc. ● Amortized Analysis ● Aggregate, banker's, and potential methods. Where We're Going ● String Data Structures ● Data structures for storing and manipulating text. ● Randomized Data Structures ● Using randomness as a building block. ● Integer Data Structures ● Breaking the Ω(n log n) sorting barrier. ● Dynamic Connectivity ● Maintaining connectivity in an changing world. String Data Structures Text Processing ● String processing shows up everywhere: ● Computational biology: Manipulating DNA sequences. ● NLP: Storing and organizing huge text databases. ● Computer security: Building antivirus databases. ● Many problems have polynomial-time solutions. ● Goal: Design theoretically and practically efficient algorithms that outperform brute-force approaches. Outline for Today ● Tries ● A fundamental building block in string processing algorithms. ● Aho-Corasick String Matching ● A fast and elegant algorithm for searching large texts for known substrings. Tries Ordered Dictionaries ● Suppose we want to store a set of elements supporting the following operations: ● Insertion of new elements. ● Deletion of old elements. ● Membership queries. ● Successor queries. ● Predecessor queries. ● Min/max queries. ● Can use a standard red/black tree or splay tree to get (worst-case or expected) O(log n) implementations of each. A Catch ● Suppose we want to store a set of strings. ● Comparing two strings of lengths r and s takes time O(min{r, s}). ● Operations on a balanced BST or splay tree now take time O(M log n), where M is the length of the longest string in the tree.